
Model Building Report
This document lists the results for the homology modelling project "nsp7 / nsp8 / Pol hetero-oligomeric complex | P0DTD1 PRO_0000449625, PRO_0000449626, PRO_0000449629" submitted to SWISS-MODEL workspace on May 5, 2023, 9:33 p.m..The submitted primary amino acid sequence is given in Table T1.
If you use any results in your research, please cite the relevant publications:
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede TSWISS-MODEL: homology modelling of protein structures and complexes.Nucleic Acids Res 46, W296-W303. (2018)
 29788355
29788355 10.1093/nar/gky427
10.1093/nar/gky427 - Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede TThe SWISS-MODEL Repository - new features and functionality.Nucleic Acids Res 45, D313-D319. (2017)
 27899672
27899672 10.1093/nar/gkw1132
10.1093/nar/gkw1132 - Studer G, Tauriello G, Bienert S, Biasini M, Johner N, Schwede TProMod3 - A versatile homology modelling toolbox.PLOS Comp Biol 17(1), e1008667. (2021)
 33507980
33507980 10.1371/journal.pcbi.1008667
10.1371/journal.pcbi.1008667 - Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede TQMEANDisCo - distance constraints applied on model quality estimation.Bioinformatics 36, 1765-1771. (2020)
 31697312
31697312 10.1093/bioinformatics/btz828
10.1093/bioinformatics/btz828 - Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede TModeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology.Scientific Reports 7. (2017)
 28874689
28874689 10.1038/s41598-017-09654-8
10.1038/s41598-017-09654-8
Results
The SWISS-MODEL template library (SMTL version 2023-05-05, PDB release 2023-04-28) was searched with for evolutionary related structures matching the target sequences in Table T1. For details on the template search, see Materials and Methods. Overall 98 templates were found (Table T2).
Models
The following model was built (see Materials and Methods "Model Building"):
Model #02 |
File | Built with | Oligo-State | Ligands | GMQE | QMEANDisCo Global |
|---|---|---|---|---|---|---|
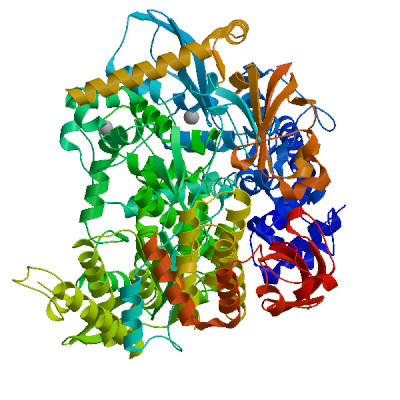
|
PDB | ProMod3 3.3.0 | hetero-1-1-2-mer |
2 x ZN: ZINC ION;
|
0.74 | 0.86 ± 0.05 |
|
|
| Template | Seq Identity | Oligo-state | QSQE | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| 6nur.1 | 96.70 | hetero-1-2-1-mer | - | HHblits | EM | - | 0.61 | C: 2-71 A: 117-919 BD: 77-191 | 1.00 | NSP7 NSP12 NSP8 |
Included Ligands
| Ligand | Description |
|---|---|
| 2 x ZN | ZINC ION |
Target SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLLSMQGAVDINKLCEEMLDNRA
6nur.1.C SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLLSMQGAVDINRLCEEMLDNRA
Target TLQ
6nur.1.C TLQ
Target SADAQSFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNDKVAGFAKFLKTNCCRFQEKDEDDNLIDSYFVVKRHTFSNY
6nur.1.A SADASTFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNEKVAGFAKFLKTNCCRFQEKDEEGNLLDSYFVVKRHTMSNY
Target QHEETIYNLLKDCPAVAKHDFFKFRIDGDMVPHISRQRLTKYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKK
6nur.1.A QHEETIYNLVKDCPAVAVHDFFKFRVDGDMVPHISRQRLTKYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKK
Target DWYDFVENPDILRVYANLGERVRQALLKTVQFCDAMRNAGIVGVLTLDNQDLNGNWYDFGDFIQTTPGSGVPVVDSYYSL
6nur.1.A DWYDFVENPDILRVYANLGERVRQSLLKTVQFCDAMRDAGIVGVLTLDNQDLNGNWYDFGDFVQVAPGCGVPIVDSYYSL
Target LMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERLKLFDRYFKYWDQTYHPNCVNCLDDRCILHCANFNVLFSTV
6nur.1.A LMPILTLTRALAAESHMDADLAKPLIKWDLLKYDFTEERLCLFDRYFKYWDQTYHPNCINCLDDRCILHCANFNVLFSTV
Target FPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVNLHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAA
6nur.1.A FPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVNLHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAA
Target LTNNVAFQTVKPGNFNKDFYDFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYF
6nur.1.A LTNNVAFQTVKPGNFNKDFYDFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYF
Target DCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTITQMNLKYAISAKNRARTVAGV
6nur.1.A DCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTITQMNLKYAISAKNRARTVAGV
Target SICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHNMLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLAR
6nur.1.A SICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHNMLKTVYSDVETPHLMGWDYPKCDRAMPNMLRIMASLVLAR
Target KHTTCCSLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYV
6nur.1.A KHNTCCNLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYV
Target RNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSDDAVVCFNSTYASQGLVASIKNFKSVLYYQNNVFMSEAKCW
6nur.1.A RNLQHRLYECLYRNRDVDHEFVDEFYAYLRKHFSMMILSDDAVVCYNSNYAAQGLVASIKNFKAVLYYQNNVFMSEAKCW
Target TETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGAGCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADV
6nur.1.A TETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGAGCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADV
Target FHLYLQYIRKLHDELTGHMLDMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQ
6nur.1.A FHLYLQYIRKLHDELTGHMLDMYSVMLTNDNTSRYWEPEFYEAMYTPHTVL-
Target AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKR
6nur.1.B AIASEFSSLPSYAAYATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKR
Target AKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVV
6nur.1.B AKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVVPDYGTYKNTCDGNTFTYASALWEIQQVV
Target DADSKIVQLSEISMDNSPNLAWPLIVTALRANSAVKLQ
6nur.1.B DADSKIVQLSEINMDNSPNLAWPLIVTALRANSAVKLQ
Target AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKR
6nur.1.D AIASEFSSLPSYAAYATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKR
Target AKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVV
6nur.1.D AKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVVPDYGTYKNTCDGNTFTYASALWEIQQVV
Target DADSKIVQLSEISMDNSPNLAWPLIVTALRANSAVKLQ
6nur.1.D DADSKIVQLSEINMDNSPNLAWPLIVTALRANSAVKLQ
Materials and Methods
Model Building
Models are built based on the target-template alignment using ProMod3 (Studer et al.). Coordinates which are conserved between the target and the template are copied from the template to the model. Insertions and deletions are remodelled using a fragment library. Side chains are then rebuilt. Finally, the geometry of the resulting model is regularized by using a force field.
Model Quality Estimation
The global and per-residue model quality has been assessed using the QMEAN scoring function (Studer et al.).
Ligand Modelling
Ligands present in the template structure are transferred by homology to the model when the following criteria are met: (a) The ligands are annotated as biologically relevant in the template library, (b) the ligand is in contact with the model, (c) the ligand is not clashing with the protein, (d) the residues in contact with the ligand are conserved between the target and the template. If any of these four criteria is not satisfied, a certain ligand will not be included in the model. The model summary includes information on why and which ligand has not been included.
Oligomeric State Conservation
The quaternary structure annotation of the template is used to model the target sequence in its oligomeric form. The method (Bertoni et al.) is based on a supervised machine learning algorithm, Support Vector Machines (SVM), which combines interface conservation, structural clustering, and other template features to provide a quaternary structure quality estimate (QSQE). The QSQE score is a number between 0 and 1, reflecting the expected accuracy of the interchain contacts for a model built based a given alignment and template. Higher numbers indicate higher reliability. This complements the GMQE score which estimates the accuracy of the tertiary structure of the resulting model.
References
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TLBLAST+: architecture and applications.BMC Bioinformatics, 10, 421-430. (2009)
 20003500
20003500 10.1186/1471-2105-10-421
10.1186/1471-2105-10-421 - Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger SJ, Söding JHH-suite3 for fast remote homology detection and deep protein annotation.BMC Bioinformatics 20, 473. (2019)
 31521110
31521110 10.1186/s12859-019-3019-7
10.1186/s12859-019-3019-7
Table T1:
Primary amino acid sequences for which templates were searched and models were built.
FFKFRIDGDMVPHISRQRLTKYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKKDWYDFVENPDILRVYANLGERVRQALLKTVQFCDAMRNAG
IVGVLTLDNQDLNGNWYDFGDFIQTTPGSGVPVVDSYYSLLMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERLKLFDRYFKYWDQTYHPNCVN
CLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVNLHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAA
LTNNVAFQTVKPGNFNKDFYDFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYFDCYDGGCINANQVIVNNLDK
SAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTITQMNLKYAISAKNRARTVAGVSICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHN
MLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLARKHTTCCSLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAV
TANVNALLSTDGNKIADKYVRNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSDDAVVCFNSTYASQGLVASIKNFKSVLYYQNNVFMSEAKCW
TETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGAGCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADVFHLYLQYIRKLHDELTGHML
DMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQ
DALNNIINNARDGCVPLNIIPLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNLAWPLIVTALRANSAVKLQ
Table T2:
| Template | Seq Identity | Oligo-state | QSQE | Found by | Method | Resolution | Seq Similarity | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 7ed5.1 | 100.00 | hetero-1-1-2-mer | 0.85 | BLAST / HHblits | EM | 2.98Å | 0.62 | 1.00 | Non-structural protein 7; RNA-directed RNA polymerase; Non-structural protein 8 |
| 7aap.1 | 100.00 | hetero-1-1-2-mer | 0.68 | HHblits | EM | NA | 0.62 | 1.00 | Non-structural protein 7; Non-structural protein 12; Non-structural protein 8 |
| 8gwe.1 | 100.00 | hetero-1-1-2-mer | 0.90 | HHblits | EM | NA | 0.62 | 1.00 | Non-structural protein 7; RNA-directed RNA polymerase; Non-structural protein 8 |
The table above shows the top 3 filtered templates. A further 55 templates were found which were considered to be less suitable for modelling than the filtered list.
6m71.1, 6nur.1, 6xez.1, 6xqb.1, 6yyt.1, 7aap.1, 7b3b.1, 7b3c.1, 7b3d.1, 7btf.1, 7bv1.1, 7bv2.1, 7bw4.1, 7bzf.1, 7c2k.1, 7ctt.1, 7cxm.1, 7cxn.1, 7cyq.1, 7d4f.1, 7dfg.1, 7dfh.1, 7doi.1, 7dok.1, 7dte.1, 7ed5.1, 7egq.1, 7eiz.1, 7krn.1, 7kro.1, 7krp.1, 7l1f.1, 7oyg.1, 7ozu.1, 7ozv.1, 7rdx.1, 7rdy.1, 7rdz.1, 7re0.1, 7re1.1, 7re2.1, 7re3.1, 7thm.1, 7uo4.1, 7uo7.1, 7uo9.1, 7uob.1, 7uoe.1, 8g6r.1, 8gwb.1, 8gwf.1, 8gwg.1, 8gwi.1, 8gwn.1, 8gwo.1