Projects created before 2024-01-08 are available to browse. Unfortunately templates originating from AlphaFold Database (AFDB) will fail for model building.
Please start a new template search to build models based on AFDB templates.
Proofreading exoribonuclease nsp14 (ExoN) | P0DTD1 PRO_0000449631
Created: May 5, 2023 at 21:33
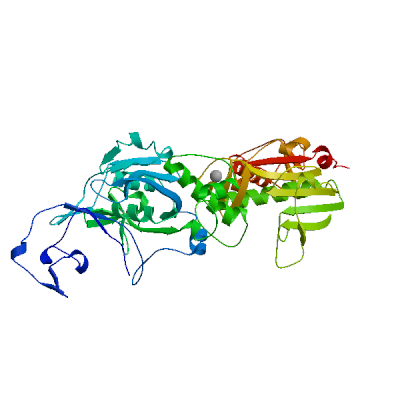
Model 01
- Ligands
2 x ZN 2 x ZINC IONZN.1: 5 residues within 4Å:- Chain A: C.207, C.208, C.210, C.226, H.229
4 PLIP interactions:4 interactions with chain A- Metal complexes: A:C.207, A:C.210, A:C.226, A:H.229
ZN.2: 4 residues within 4Å:- Chain A: H.257, C.261, H.264, C.279
4 PLIP interactions:4 interactions with chain A- Metal complexes: A:H.257, A:C.261, A:H.264, A:C.279
QMEANDisCo Local
QMEAN Z-Scores
Template
5nfy.1.A
Polyprotein 1ab
SARS-CoV nsp10/nsp14 dynamic complex
SARS-CoV nsp10/nsp14 dynamic complex
- Seq Identity
- 94.88%
- Coverage
| Biounit Oligo State | Hetero-dimer |
| QSQE | 0.00 |
| Method | X-ray, 3.38 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 3-525 |
Model-Template Alignment
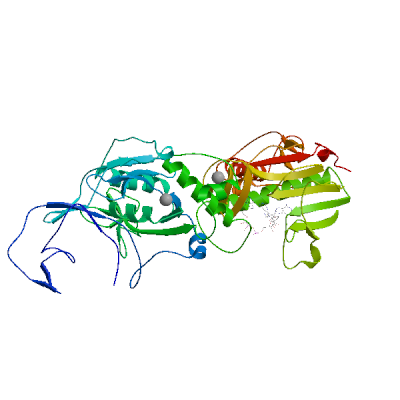
Model 02
- Ligands
1 x G3A ,1 x MG ,1 x SAH ,1 x ZN 1 x GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATEG3A.8: 17 residues within 4Å:- Chain B: N.306, C.309, R.310, Q.313, K.336, I.338, W.385, N.386, N.388, F.401, Y.420, N.422, K.423, F.426, T.428, F.506,
- Ligand: SAH.7
15 PLIP interactions:15 interactions with chain B- Hydrogen bonds: B:N.306, B:N.306, B:R.310, B:R.310, B:I.338, B:N.386, B:N.388, B:N.388
- Salt bridges: B:R.310, B:R.310, B:R.310
- pi-Stacking: B:W.385, B:F.426
- pi-Cation interactions: B:R.310, B:R.310
1 x MAGNESIUM IONMG.6: 3 residues within 4Å:- Chain B: D.90, A.187, E.191
3 PLIP interactions:3 interactions with chain B- Metal complexes: B:D.90, B:E.191, B:E.191
1 x S-ADENOSYL-L-HOMOCYSTEINESAH.7: 14 residues within 4Å:- Chain B: W.292, G.333, N.334, P.335, D.352, A.353, Q.354, L.366, F.367, Y.368, W.385, N.386, C.387, V.389
5 PLIP interactions:5 interactions with chain B- Hydrogen bonds: B:A.353, B:Q.354, B:Y.368, B:Y.368, B:W.385
1 x ZINC IONZN.4: 4 residues within 4Å:- Chain B: H.257, C.261, H.264, C.279
4 PLIP interactions:4 interactions with chain B- Metal complexes: B:H.257, B:C.261, B:H.264, B:C.279
QMEANDisCo Local
QMEAN Z-Scores
Template
5c8s.1.B
Guanine-N7 methyltransferase
Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA
Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA
- Seq Identity
- 95.07%
- Coverage
| Biounit Oligo State | Hetero-dimer |
| QSQE | 0.00 |
| Method | X-ray, 3.33 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 1-525 |
Model-Template Alignment
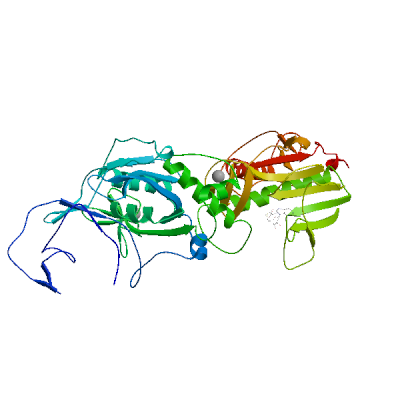
Model 03
- Ligands
1 x SAM ,2 x ZN 1 x S-ADENOSYLMETHIONINESAM.7: 13 residues within 4Å:- Chain B: W.292, G.333, P.335, D.352, A.353, Q.354, L.366, F.367, Y.368, W.385, N.386, C.387, V.389
8 PLIP interactions:8 interactions with chain B- Hydrophobic interactions: B:N.386
- Hydrogen bonds: B:G.333, B:A.353, B:Q.354, B:Y.368, B:Y.368, B:W.385
- Salt bridges: B:R.310
2 x ZINC IONZN.3: 4 residues within 4Å:- Chain B: C.207, C.210, C.226, H.229
4 PLIP interactions:4 interactions with chain B- Metal complexes: B:C.210, B:C.226, B:H.229, B:H.229
ZN.4: 4 residues within 4Å:- Chain B: H.257, C.261, H.264, C.279
3 PLIP interactions:3 interactions with chain B- Metal complexes: B:H.257, B:C.261, B:H.264
QMEANDisCo Local
QMEAN Z-Scores
Template
5c8t.1.B
Guanine-N7 methyltransferase
Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM
Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM
- Seq Identity
- 95.07%
- Coverage
| Biounit Oligo State | Hetero-dimer |
| QSQE | 0.00 |
| Method | X-ray, 3.20 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 1-525 |
Model-Template Alignment
Delete Model - ""
Are you sure you want to delete this model?
(This really can't be undone!)
Delete Project - ""
Are you sure you want to delete this project?
(This really can't be undone!)