Projects created before 2024-01-08 are available to browse. Unfortunately templates originating from AlphaFold Database (AFDB) will fail for model building.
Please start a new template search to build models based on AFDB templates.
Uridylate-specific endoribonuclease nsp15 | P0DTD1 PRO_0000449632
Created: May 5, 2023 at 21:33
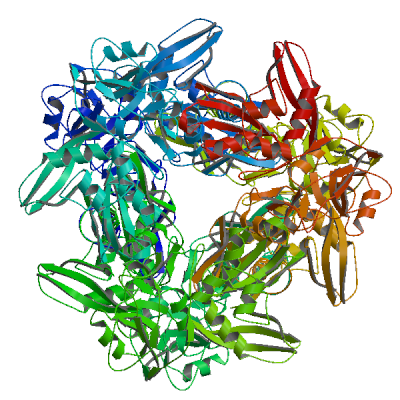
Model 04
- Ligands
6 x U5P 6 x URIDINE-5'-MONOPHOSPHATEU5P.1: 9 residues within 4Å:- Chain A: H.249, K.289, V.291, C.292, S.293, Y.342, P.343, K.344, L.345
5 PLIP interactions:5 interactions with chain A- Hydrogen bonds: A:H.249, A:S.293, A:S.293, A:S.293, A:L.345
U5P.11: 8 residues within 4Å:- Chain B: H.249, K.289, V.291, C.292, S.293, Y.342, K.344, L.345
6 PLIP interactions:6 interactions with chain B- Hydrophobic interactions: B:Y.342
- Hydrogen bonds: B:H.249, B:S.293, B:S.293, B:S.293, B:L.345
U5P.22: 9 residues within 4Å:- Chain C: H.249, K.289, V.291, C.292, S.293, Y.342, P.343, K.344, L.345
5 PLIP interactions:5 interactions with chain C- Hydrogen bonds: C:H.249, C:S.293, C:S.293, C:S.293, C:L.345
U5P.32: 8 residues within 4Å:- Chain D: H.249, K.289, V.291, C.292, S.293, Y.342, K.344, L.345
6 PLIP interactions:6 interactions with chain D- Hydrophobic interactions: D:Y.342
- Hydrogen bonds: D:H.249, D:S.293, D:S.293, D:S.293, D:L.345
U5P.43: 9 residues within 4Å:- Chain E: H.249, K.289, V.291, C.292, S.293, Y.342, P.343, K.344, L.345
5 PLIP interactions:5 interactions with chain E- Hydrogen bonds: E:H.249, E:S.293, E:S.293, E:S.293, E:L.345
U5P.53: 8 residues within 4Å:- Chain F: H.249, K.289, V.291, C.292, S.293, Y.342, K.344, L.345
6 PLIP interactions:6 interactions with chain F- Hydrophobic interactions: F:Y.342
- Hydrogen bonds: F:H.249, F:S.293, F:S.293, F:S.293, F:L.345
QMEANDisCo Local
QMEAN Z-Scores
Template
6wlc.1.A
Uridylate-specific endoribonuclease
Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate
Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Homo-hexamer |
| QSQE | 0.92 |
| Method | X-ray, 1.82 Å |
| Seq Similarity | 0.61 |
| Coverage | 1.00 |
| Range | 1-346 |
| Ligand | Added to Model | Description |
|---|---|---|
| ✓ | URIDINE-5'-MONOPHOSPHATE | |
| ✓ | URIDINE-5'-MONOPHOSPHATE | |
| ✓ | URIDINE-5'-MONOPHOSPHATE | |
| ✓ | URIDINE-5'-MONOPHOSPHATE | |
| ✓ | URIDINE-5'-MONOPHOSPHATE | |
| ✓ | URIDINE-5'-MONOPHOSPHATE | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | ACETATE ION | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | FORMIC ACID | |
| ✕ - Not in contact with model. | FORMIC ACID | |
| ✕ - Not biologically relevant. | FORMIC ACID | |
| ✕ - Not in contact with model. | FORMIC ACID | |
| ✕ - Not biologically relevant. | FORMIC ACID | |
| ✕ - Not in contact with model. | FORMIC ACID | |
| ✕ - Not biologically relevant. | SULFATE ION | |
| ✕ - Not biologically relevant. | SULFATE ION | |
| ✕ - Not biologically relevant. | SULFATE ION | |
| ✕ - Not biologically relevant. | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | |
| ✕ - Not biologically relevant. | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | |
| ✕ - Not biologically relevant. | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | |
| ✕ - Not biologically relevant. | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | |
| ✕ - Not biologically relevant. | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | |
| ✕ - Not biologically relevant. | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL |
Model-Template Alignment
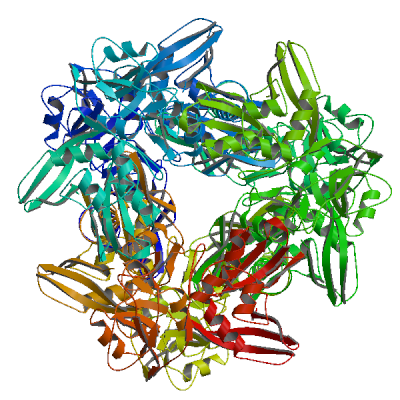
Model 02
QMEANDisCo Local
QMEAN Z-Scores
Template
6w01.1.A
Uridylate-specific endoribonuclease
The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate
The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Homo-hexamer |
| QSQE | 0.92 |
| Method | X-ray, 1.90 Å |
| Seq Similarity | 0.61 |
| Coverage | 1.00 |
| Range | 1-345 |
| Ligand | Added to Model | Description |
|---|---|---|
| ✕ - Not biologically relevant. | CITRIC ACID | |
| ✕ - Not biologically relevant. | CITRIC ACID | |
| ✕ - Not biologically relevant. | CITRIC ACID | |
| ✕ - Not biologically relevant. | CITRIC ACID | |
| ✕ - Not biologically relevant. | CITRIC ACID | |
| ✕ - Not biologically relevant. | CITRIC ACID | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not in contact with model. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | 1,2-ETHANEDIOL | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER | |
| ✕ - Not biologically relevant. | DI(HYDROXYETHYL)ETHER |
Model-Template Alignment
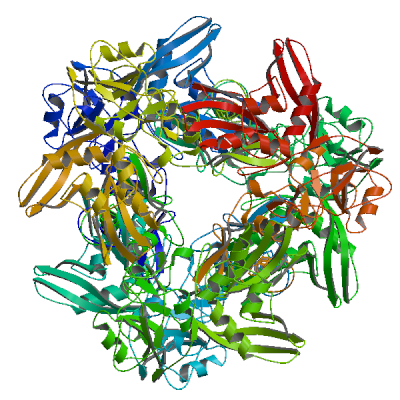
Model 01
QMEANDisCo Local
QMEAN Z-Scores
Template
2h85.1.A
Putative orf1ab polyprotein
Crystal Structure of Nsp 15 from SARS
Crystal Structure of Nsp 15 from SARS
- Seq Identity
- 88.12%
- Coverage
| Biounit Oligo State | Homo-hexamer |
| QSQE | 0.87 |
| Method | X-ray, 2.60 Å |
| Seq Similarity | 0.58 |
| Coverage | 1.00 |
| Range | 1-345 |
Model-Template Alignment
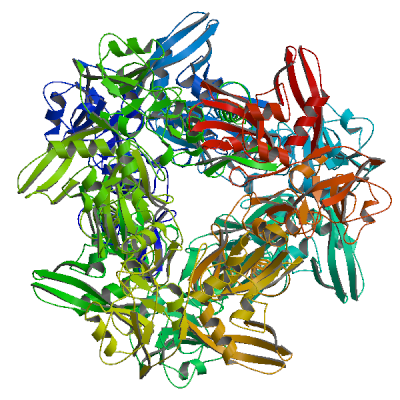
Model 03
QMEANDisCo Local
QMEAN Z-Scores
Template
2rhb.1.A
Uridylate-specific endoribonuclease
Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit
Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit
- Seq Identity
- 88.44%
- Coverage
| Biounit Oligo State | Homo-hexamer |
| QSQE | 0.88 |
| Method | X-ray, 2.80 Å |
| Seq Similarity | 0.58 |
| Coverage | 1.00 |
| Range | 1-344 |
Model-Template Alignment
Delete Model - ""
Are you sure you want to delete this model?
(This really can't be undone!)
Delete Project - ""
Are you sure you want to delete this project?
(This really can't be undone!)