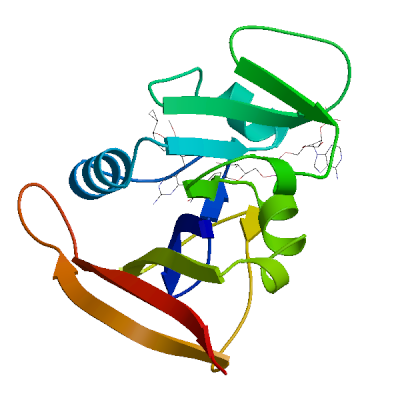
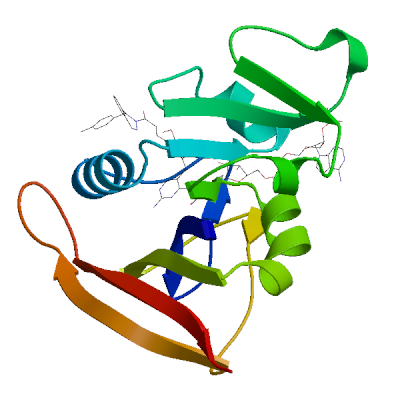
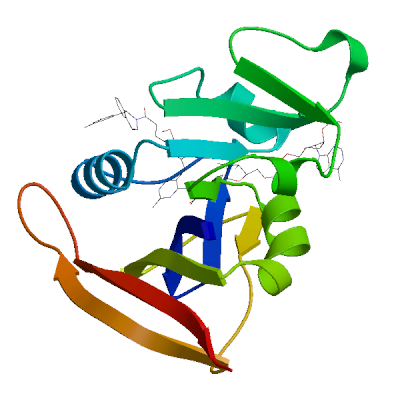
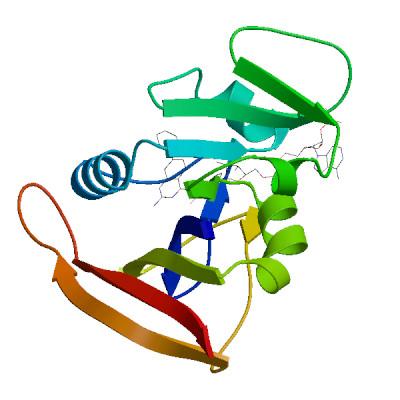
Model 01
- Ligands
1 x MMV ,1 x NAP 1 x 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acidMMV.2: 13 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, D.28, L.29, V.32, T.47, S.50, I.51, F.93, T.112,
- Ligand: NAP.1
9 PLIP interactions:9 interactions with chain A- Hydrophobic interactions: A:L.21, A:L.21, A:T.47, A:I.51, A:F.93
- Hydrogen bonds: A:L.6, A:T.112, A:T.112
- Salt bridges: A:D.28
1 x NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENAP.1: 27 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, F.17, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, L.98, F.99, E.101, T.122
20 PLIP interactions:20 interactions with chain A- Hydrophobic interactions: A:T.47, A:F.99
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:G.44, A:R.45, A:R.45, A:K.46, A:T.47, A:T.47, A:T.64, A:S.65, A:G.95, A:Q.96, A:T.97, A:T.97, A:E.101
- Salt bridges: A:R.45
QMEANDisCo Local
QMEAN Z-Scores
Template
6e4e.1.A
Dihydrofolate reductase
Crystal structure of dihydrofolate reductase from Staphylococcus aureus MW2 bound to NADP and p218
Crystal structure of dihydrofolate reductase from Staphylococcus aureus MW2 bound to NADP and p218
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 1.90 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 1-159 |
Model-Template Alignment
Model 05
- Ligands
1 x NDP ,1 x XCF 1 x NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENDP.1: 25 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, L.98, E.101, T.122
20 PLIP interactions:20 interactions with chain A- Hydrophobic interactions: A:T.47
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:R.45, A:R.45, A:K.46, A:T.47, A:T.64, A:S.65, A:D.66, A:G.95, A:Q.96, A:T.97, A:L.98, A:E.101, A:T.122
- Salt bridges: A:R.45, A:K.46
1 x 5-[[(2R)-2-cyclopropyl-7,8-dimethoxy-2H-chromen-5-yl]methyl]pyrimidine-2,4-diamineXCF.2: 11 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, D.28, L.29, V.32, S.50, L.55, F.93,
- Ligand: NDP.1
10 PLIP interactions:10 interactions with chain A- Hydrophobic interactions: A:L.21, A:L.29, A:L.55, A:F.93, A:F.93
- Hydrogen bonds: A:L.6, A:V.7, A:A.8, A:L.21
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
3fyw.1.A
Dihydrofolate reductase
Staph. aureus DHFR complexed with NADPH and AR-101
Staph. aureus DHFR complexed with NADPH and AR-101
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 2.10 Å |
| Seq Similarity | 0.62 |
| Coverage | 0.99 |
| Range | 2-159 |
Model-Template Alignment
Model 03
- Ligands
1 x NAP ,1 x OWS 1 x NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENAP.1: 26 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, F.17, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, L.98, E.101, T.122
24 PLIP interactions:24 interactions with chain A- Hydrophobic interactions: A:L.21, A:T.47
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:G.44, A:R.45, A:R.45, A:K.46, A:T.47, A:T.47, A:T.64, A:T.64, A:S.65, A:G.95, A:Q.96, A:T.97, A:T.97, A:T.97, A:L.98, A:E.101
- Salt bridges: A:R.45, A:K.46
1 x (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(4-methylphenyl)phthalazin-2(1H)-yl]prop-2-en-1-oneOWS.2: 18 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, P.26, D.28, L.29, K.30, V.32, K.33, S.50, I.51, L.55, P.56, R.58, F.93, T.112,
- Ligand: NAP.1
17 PLIP interactions:17 interactions with chain A- Hydrophobic interactions: A:L.21, A:L.29, A:L.29, A:L.29, A:K.30, A:K.33, A:K.33, A:L.55, A:P.56, A:F.93, A:F.93
- Hydrogen bonds: A:L.6, A:V.7, A:A.8, A:S.50, A:T.112
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
6pr6.1.A
Dihydrofolate reductase
S. aureus dihydrofolate reductase co-crystallized with para-tolyl-dihydropthalazine inhibitor and NADP(H)
S. aureus dihydrofolate reductase co-crystallized with para-tolyl-dihydropthalazine inhibitor and NADP(H)
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 2.01 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 2-159 |
| Ligand | Added to Model | Description |
|---|---|---|
| ✓ | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | |
| ✓ | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(4-methylphenyl)phthalazin-2(1H)-yl]prop-2-en-1-one | |
| ✕ - Not biologically relevant. | GLYCEROL | |
| ✕ - Not biologically relevant. | GLYCEROL |
Model-Template Alignment
Model 06
- Ligands
1 x NAP ,1 x OWJ 1 x NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENAP.2: 26 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, L.98, E.101, T.122,
- Ligand: OWJ.1
24 PLIP interactions:24 interactions with chain A- Hydrophobic interactions: A:L.21, A:T.47
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:R.45, A:R.45, A:K.46, A:T.47, A:T.47, A:T.64, A:T.64, A:S.65, A:G.94, A:G.95, A:Q.96, A:T.97, A:T.97, A:T.97, A:L.98, A:E.101
- Salt bridges: A:R.45, A:K.46
1 x (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(3,5-dimethylphenyl)phthalazin-2(1H)-yl]prop-2-en-1-oneOWJ.1: 16 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, D.28, L.29, K.30, V.32, K.33, S.50, I.51, L.55, P.56, R.58, F.93, T.112
14 PLIP interactions:14 interactions with chain A- Hydrophobic interactions: A:L.21, A:L.29, A:L.29, A:K.30, A:K.33, A:L.55, A:P.56, A:F.93, A:F.93
- Hydrogen bonds: A:L.6, A:V.7, A:S.50, A:T.112
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
6pr8.1.A
Dihydrofolate reductase
S. aureus dihydrofoate reductase co-crystallized with 3,5-dimethylphenyl-dihydropthalazine inhibitor and NADP(H)
S. aureus dihydrofoate reductase co-crystallized with 3,5-dimethylphenyl-dihydropthalazine inhibitor and NADP(H)
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 2.01 Å |
| Seq Similarity | 0.62 |
| Coverage | 0.99 |
| Range | 2-159 |
| Ligand | Added to Model | Description |
|---|---|---|
| ✓ | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | |
| ✓ | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(3,5-dimethylphenyl)phthalazin-2(1H)-yl]prop-2-en-1-one | |
| ✕ - Not biologically relevant. | GLYCEROL | |
| ✕ - Not biologically relevant. | GLYCEROL |
Model-Template Alignment
Model 04
- Ligands
1 x NAP ,1 x Q11 1 x NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENAP.1: 26 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, F.17, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, L.98, E.101, T.122
23 PLIP interactions:23 interactions with chain A- Hydrophobic interactions: A:L.21, A:T.47
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:G.44, A:R.45, A:R.45, A:K.46, A:T.47, A:T.47, A:T.64, A:T.64, A:S.65, A:G.95, A:Q.96, A:T.97, A:T.97, A:L.98, A:E.101
- Salt bridges: A:R.45, A:K.46
1 x 7-(2-methoxyphenyl)quinazoline-2,4-diamineQ11.2: 10 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, D.28, L.29, V.32, F.93, T.112,
- Ligand: NAP.1
8 PLIP interactions:8 interactions with chain A- Hydrophobic interactions: A:L.29, A:F.93, A:F.93
- Hydrogen bonds: A:L.6, A:V.7, A:A.8, A:T.112
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
3sqy.1.A
Dihydrofolate reductase
S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines
S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 1.50 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 2-158 |
Model-Template Alignment
Model 07
- Ligands
1 x NAP ,1 x Q12 1 x NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENAP.1: 26 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, F.17, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, L.98, E.101, T.122
23 PLIP interactions:23 interactions with chain A- Hydrophobic interactions: A:L.21, A:T.47
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:G.44, A:R.45, A:R.45, A:K.46, A:T.47, A:T.47, A:T.64, A:T.64, A:S.65, A:G.95, A:Q.96, A:T.97, A:T.97, A:L.98, A:E.101
- Salt bridges: A:R.45, A:K.46
1 x 7-(3,4-dimethoxyphenyl)-6-methylquinazoline-2,4-diamineQ12.2: 12 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, D.28, L.29, V.32, I.51, L.55, F.93, T.112,
- Ligand: NAP.1
8 PLIP interactions:8 interactions with chain A- Hydrophobic interactions: A:L.29, A:I.51, A:F.93, A:F.93, A:F.93
- Hydrogen bonds: A:L.6, A:V.7
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
3sr5.1.A
Dihydrofolate reductase
S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines
S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 1.68 Å |
| Seq Similarity | 0.62 |
| Coverage | 0.99 |
| Range | 2-158 |
Model-Template Alignment
Model 08
- Ligands
1 x NDP ,1 x TOP 1 x NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATENDP.2: 26 residues within 4Å:- Chain A: V.7, A.8, I.15, G.16, F.17, N.19, Q.20, L.21, W.23, G.44, R.45, K.46, T.47, L.63, T.64, S.65, H.78, I.80, F.93, G.94, G.95, Q.96, T.97, E.101, T.122,
- Ligand: TOP.1
21 PLIP interactions:21 interactions with chain A- Hydrophobic interactions: A:L.21, A:T.47
- Hydrogen bonds: A:A.8, A:I.15, A:N.19, A:N.19, A:R.45, A:R.45, A:K.46, A:T.47, A:T.64, A:S.65, A:G.95, A:Q.96, A:T.97, A:T.97, A:L.98, A:E.101, A:T.122
- Salt bridges: A:R.45, A:K.46
1 x TRIMETHOPRIMTOP.1: 11 residues within 4Å:- Chain A: L.6, V.7, A.8, L.21, D.28, L.29, V.32, S.50, I.51, F.93, T.112
7 PLIP interactions:7 interactions with chain A- Hydrophobic interactions: A:L.21, A:F.93
- Hydrogen bonds: A:L.6, A:V.7, A:S.50, A:T.112
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
2w9g.1.A
DIHYDROFOLATE REDUCTASE
Wild-type Staphylococcus aureus DHFR in complex with NADPH and trimethoprim
Wild-type Staphylococcus aureus DHFR in complex with NADPH and trimethoprim
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 1.95 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 2-158 |
Model-Template Alignment
Model 09
- Ligands
1 x TOP 1 x TRIMETHOPRIMTOP.2: 12 residues within 4Å:- Chain A: L.6, V.7, A.8, N.19, L.21, D.28, L.29, V.32, S.50, I.51, F.93, T.112
9 PLIP interactions:9 interactions with chain A- Hydrophobic interactions: A:L.21, A:F.93
- Hydrogen bonds: A:L.6, A:A.8, A:L.21, A:S.50, A:T.112, A:T.112
- Salt bridges: A:D.28
QMEANDisCo Local
QMEAN Z-Scores
Template
2w9h.1.A
DIHYDROFOLATE REDUCTASE
Wild-type Staphylococcus aureus DHFR in complex with trimethoprim
Wild-type Staphylococcus aureus DHFR in complex with trimethoprim
- Seq Identity
- 100.00%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 1.48 Å |
| Seq Similarity | 0.62 |
| Coverage | 1.00 |
| Range | 2-158 |
Model-Template Alignment
Model 02
QMEANDisCo Local
QMEAN Z-Scores
Template
2w3m.1.A
DIHYDROFOLATE REDUCTASE
HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND FOLATE
HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND FOLATE
- Seq Identity
- 30.07%
- Coverage
| Biounit Oligo State | Monomer |
| QSQE | 0.00 |
| Method | X-ray, 1.60 Å |
| Seq Similarity | 0.37 |
| Coverage | 0.90 |
| Range | 2-144 |
Model-Template Alignment
Delete Model - ""
Are you sure you want to delete this model?
(This really can't be undone!)
Delete Project - ""
Are you sure you want to delete this project?
(This really can't be undone!)