Untitled Project (Created: 4 days, 21 hours ago)Project ArchiveTotal model count : 1. | |
| Uploaded Structure: | selected_prediction.pdb |
|---|---|
| Method: | QMEANDisCo |
| QMEAN Version: | 4.5.0 |
| SMTL Version: | 2025-11-12 |
| SEQRES: | Not specified - sequence was extracted from coordinates. |
| Results: | JSON |
Quality for selected_prediction.pdb
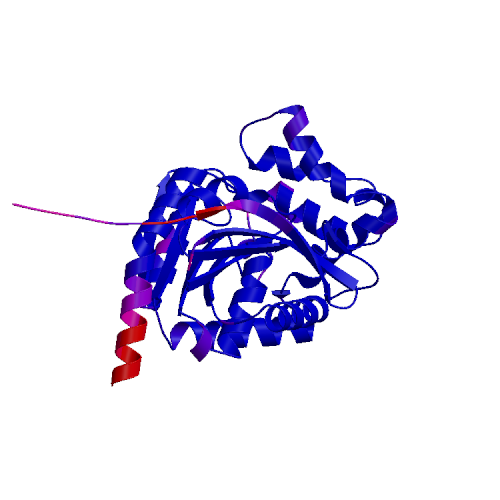
QMEANDisCo Global:0.83 ± 0.05
⚠
During preprocessing,
493 atoms were removed
.
removing atoms with zero occupancy --> removed 0 atoms with zero occupancy removing hydrogen atoms --> removed 493 hydrogen atoms
Reference for the QMEAN scoring function:
Reference for the QMEANDisCo scoring function:
Reference for the QMEANBrane scoring function:
[1]
Benkert P, Biasini M, Schwede T
Toward the estimation of the absolute quality of individual protein structure models.
Bioinformatics 27, 343-350. (2011)  21134891
21134891 10.1093/bioinformatics/btq662
10.1093/bioinformatics/btq662
 21134891
21134891 10.1093/bioinformatics/btq662
10.1093/bioinformatics/btq662Reference for the QMEANDisCo scoring function:
[2]
Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede T
QMEANDisCo - distance constraints applied on model quality estimation.
Bioinformatics 36, 1765-1771. (2020)  31697312
31697312 10.1093/bioinformatics/btz828
10.1093/bioinformatics/btz828
 31697312
31697312 10.1093/bioinformatics/btz828
10.1093/bioinformatics/btz828Reference for the QMEANBrane scoring function:
[3]
Studer G, Biasini M, Schwede T
Assessing the local structural quality of transmembrane protein models using statistical potentials (QMEANBrane).
Bioinformatics 30, i505–i511. (2014)  25161240
25161240 10.1093/bioinformatics/btu457
10.1093/bioinformatics/btu457
 25161240
25161240 10.1093/bioinformatics/btu457
10.1093/bioinformatics/btu457