Projects created before 2024-01-08 are available to browse. Unfortunately templates originating from AlphaFold Database (AFDB) will fail for model building.
Please start a new template search to build models based on AFDB templates.
Non-structural protein 8 (nsp8) | P0DTD1 PRO_0000449626
Created: May 5, 2023 at 21:34
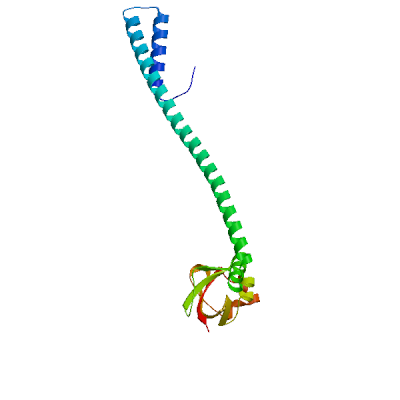
Model 01
QMEANDisCo Local
QMEAN Z-Scores
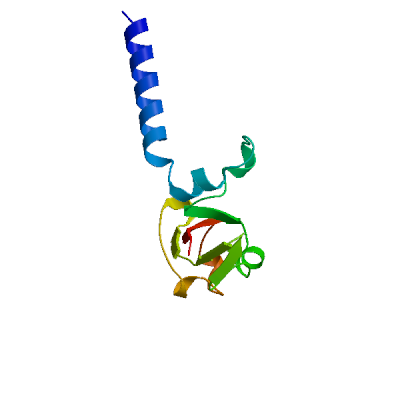
Template
2ahm.1.H
Replicase polyprotein 1ab, heavy chain
Crystal structure of SARS-CoV super complex of non-structural proteins: the hexadecamer
Crystal structure of SARS-CoV super complex of non-structural proteins: the hexadecamer
- Seq Identity
- 97.47%
- Coverage
Model-Template Alignment
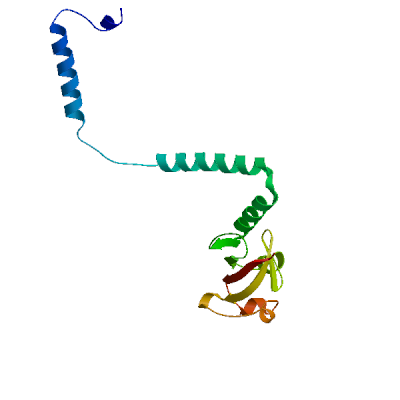
Model 04
QMEANDisCo Local
QMEAN Z-Scores
Template
2ahm.1.E
Replicase polyprotein 1ab, heavy chain
Crystal structure of SARS-CoV super complex of non-structural proteins: the hexadecamer
Crystal structure of SARS-CoV super complex of non-structural proteins: the hexadecamer
- Seq Identity
- 97.47%
- Coverage
Model-Template Alignment
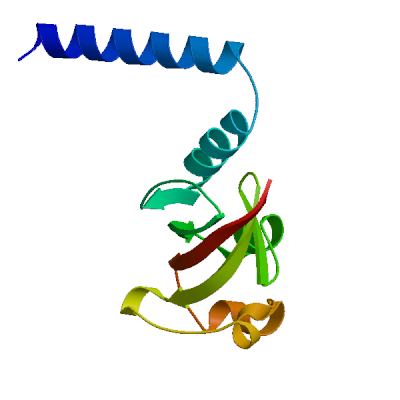
Model 05
QMEANDisCo Local
QMEAN Z-Scores
Template
7jlt.1.D
Non-structural protein 8
Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex.
Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex.
- Seq Identity
- 100.00%
- Coverage
Model-Template Alignment
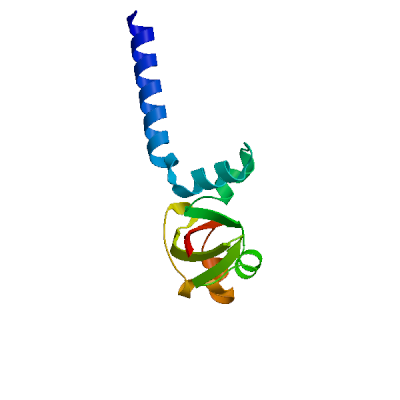
Model 02
QMEANDisCo Local
QMEAN Z-Scores
Template
6nur.1.B
NSP8
SARS-Coronavirus NSP12 bound to NSP7 and NSP8 co-factors
SARS-Coronavirus NSP12 bound to NSP7 and NSP8 co-factors
- Seq Identity
- 97.47%
- Coverage
Model-Template Alignment
Model 03
QMEANDisCo Local
QMEAN Z-Scores
Template
6m71.1.D
Non-structural protein 8
SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors
SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors
- Seq Identity
- 100.00%
- Coverage
Model-Template Alignment