Projects created before 2024-01-08 are available to browse. Unfortunately templates originating from AlphaFold Database (AFDB) will fail for model building.
Please start a new template search to build models based on AFDB templates.
Nucleoprotein (N) | P0DTC9
Created: May 5, 2023 at 21:34
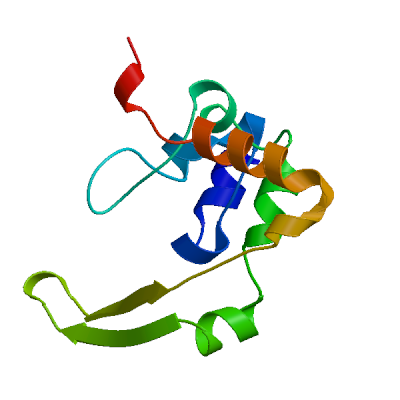
Model 02
QMEANDisCo Local
QMEAN Z-Scores
Template
4ud1.1.A
N PROTEIN
Structure of the N Terminal domain of the MERS CoV nucleocapsid
Structure of the N Terminal domain of the MERS CoV nucleocapsid
- Seq Identity
- 55.41%
- Coverage
Model-Template Alignment
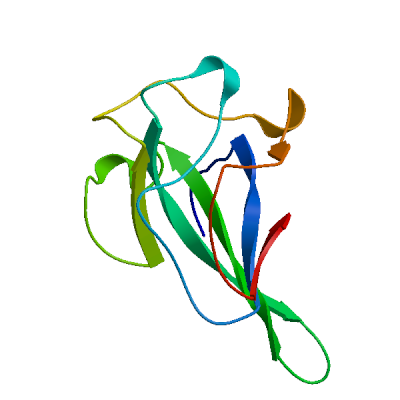
Model 01
QMEANDisCo Local
QMEAN Z-Scores
Template
2ofz.1.A
Nucleocapsid protein
Ultrahigh Resolution Crystal Structure of RNA Binding Domain of SARS Nucleopcapsid (N Protein) at 1.1 Angstrom Resolution in Monoclinic Form.
Ultrahigh Resolution Crystal Structure of RNA Binding Domain of SARS Nucleopcapsid (N Protein) at 1.1 Angstrom Resolution in Monoclinic Form.
- Seq Identity
- 92.06%
- Coverage
Model-Template Alignment
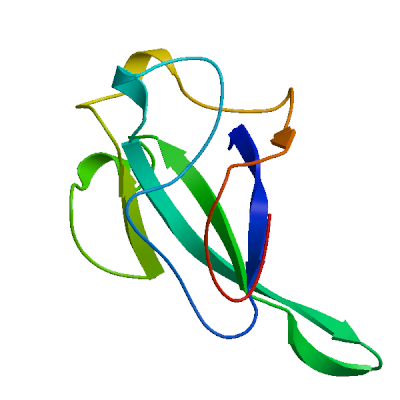
Model 03
QMEANDisCo Local
QMEAN Z-Scores
Template
6wzq.2.B
Nucleoprotein
Structure of SARS-CoV-2 Nucleocapsid dimerization domain, P21 form
Structure of SARS-CoV-2 Nucleocapsid dimerization domain, P21 form
- Seq Identity
- 100.00%
- Coverage
Model-Template Alignment