Projects created before 2024-01-08 are available to browse. Unfortunately templates originating from AlphaFold Database (AFDB) will fail for model building.
Please start a new template search to build models based on AFDB templates.
3C-like proteinase nsp5 (3CL-PRO) | P0DTD1 PRO_0000449623
Created: May 5, 2023 at 21:34
Project Summary
Template Results
A total of 2024 templates were found to match the target sequence. This list was filtered by a heuristic down to 14. The top templates are:| Template | Sequence Identity | Biounit Oligo State | Description |
|---|---|---|---|
| 6lu7.1 | 100.00 | homo-dimer | 3C-like proteinase The crystal structure of COVID-19 main protease in complex with an inhibitor N3 |
| 6y2g.1 | 100.00 | homo-dimer | 3C-like proteinase Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) |
| 5ree.1 | 100.00 | homo-dimer | 3C-like proteinase PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2217052426 |
| 5rea.1 | 100.00 | homo-dimer | 3C-like proteinase PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z31432226 |
| 5r7z.1 | 100.00 | homo-dimer | 3C-like proteinase PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1220452176 |
Model Results
The top models are:| Id | Template | GMQE | QMEANDisCo Global | Oligo State | Ligands | |
|---|---|---|---|---|---|---|
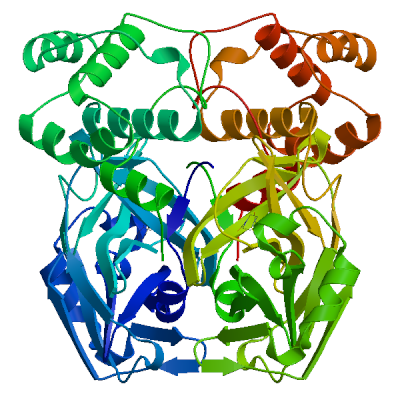 | 02 | 5rfv.1.B | 0.97 | 0.91 ± 0.05 | homo-dimer | 2 x T8J |
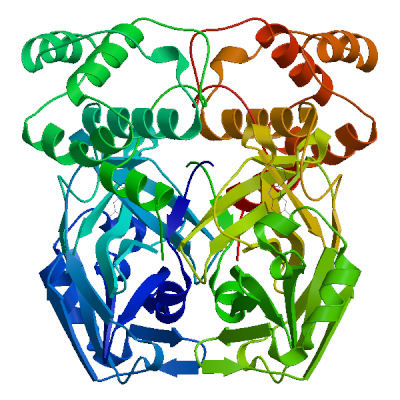 | 04 | 5r7z.1.B | 0.97 | 0.91 ± 0.05 | homo-dimer | 2 x HWH |
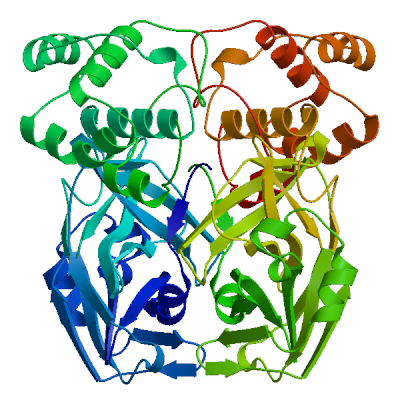 | 05 | 6lu7.1.B | 0.97 | 0.89 ± 0.05 | homo-dimer | - |
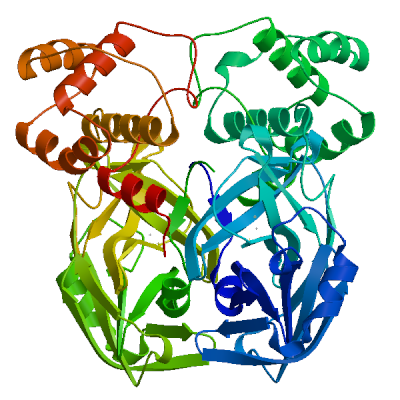 | 06 | 2z9j.1.A | 0.96 | 0.89 ± 0.05 | homo-dimer | 2 x DTZ |
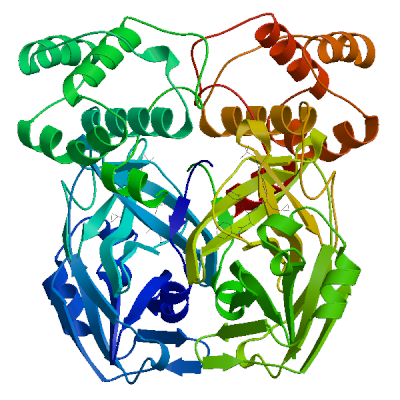 | 01 | 6y2g.1.A | 0.95 | 0.88 ± 0.05 | homo-dimer | 1 x GLY, 2 x O6K |
