Projects created before 2024-01-08 are available to browse. Unfortunately templates originating from AlphaFold Database (AFDB) will fail for model building.
Please start a new template search to build models based on AFDB templates.
Papain-like protease nsp3 | P0DTD1 PRO_0000449621
Created: May 5, 2023 at 21:33
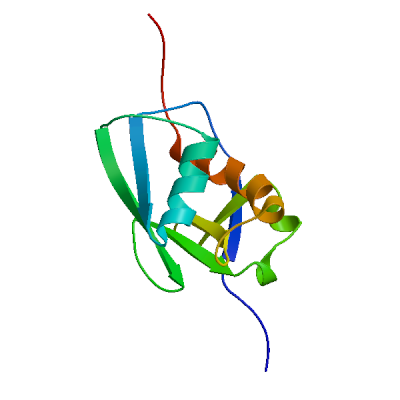
Model 10
QMEANDisCo Local
QMEAN Z-Scores
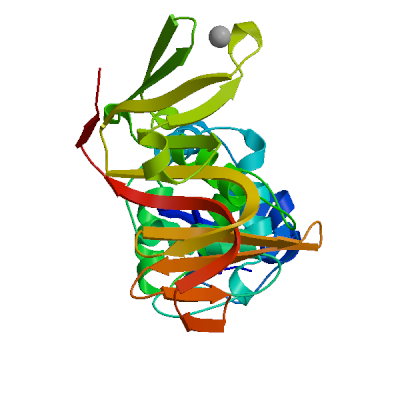
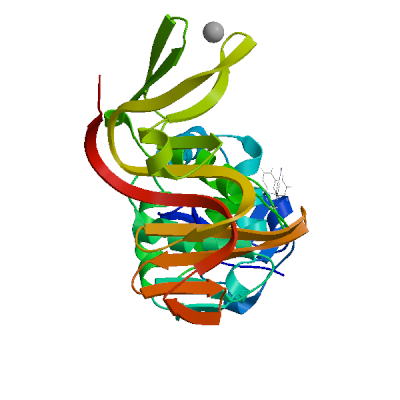
Template
3e9s.1.A
Non-structural protein 3
A new class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
A new class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
- Seq Identity
- 82.86%
- Coverage
Model-Template Alignment
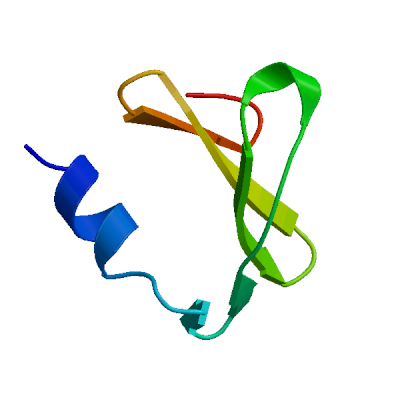
Model 01
QMEANDisCo Local
QMEAN Z-Scores
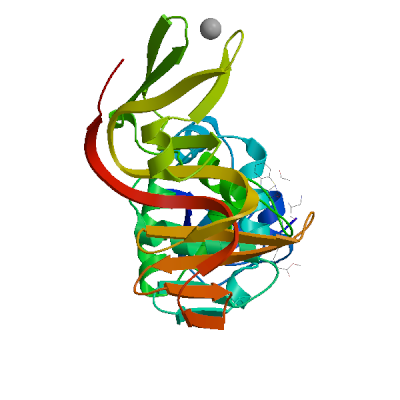
Template
3e9s.1.A
Non-structural protein 3
A new class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
A new class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
- Seq Identity
- 82.86%
- Coverage
Model-Template Alignment
Model 09
QMEANDisCo Local
QMEAN Z-Scores
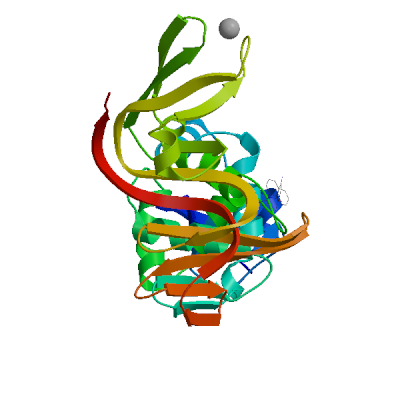
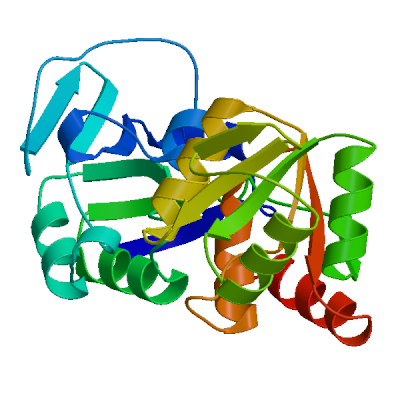
Template
7cmd.4.A
Non-structural protein 3
Crystal structure of the SARS-CoV-2 PLpro with GRL0617
Crystal structure of the SARS-CoV-2 PLpro with GRL0617
- Seq Identity
- 100.00%
- Coverage
Model-Template Alignment
Model 08
QMEANDisCo Local
QMEAN Z-Scores
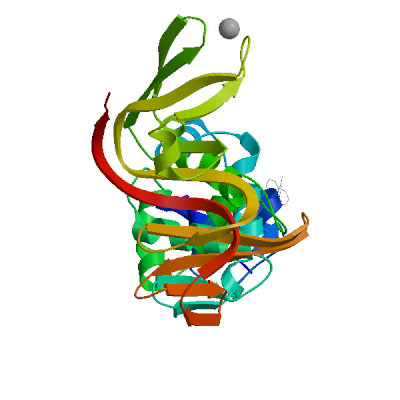
Template
2w2g.1.A
NON-STRUCTURAL PROTEIN 3
HUMAN SARS CORONAVIRUS UNIQUE DOMAIN
HUMAN SARS CORONAVIRUS UNIQUE DOMAIN
- Seq Identity
- 75.00%
- Coverage
Model-Template Alignment
Model 05
QMEANDisCo Local
QMEAN Z-Scores
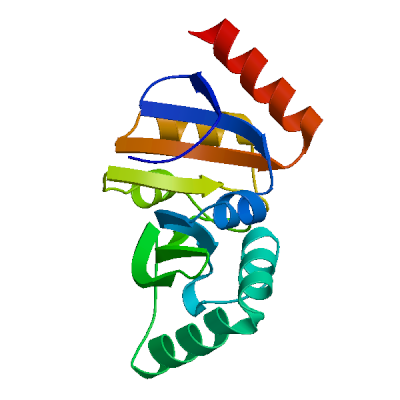
Template
6ywk.3.A
NSP3 macrodomain
Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES
Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES
- Seq Identity
- 100.00%
- Coverage
Model-Template Alignment
Model 06
QMEANDisCo Local
QMEAN Z-Scores
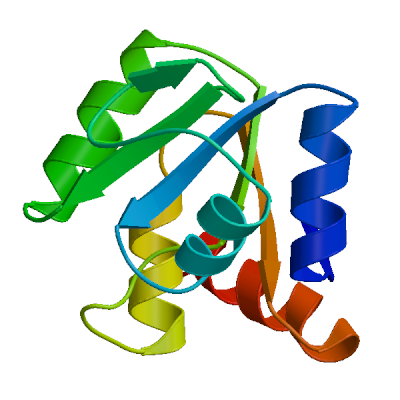
Template
2kqv.1.A
Non-structural protein 3
SARS coronavirus-unique domain (SUD): Three-domain molecular architecture in solution and RNA binding. I: Structure of the SUD-M domain of SUD-MC
SARS coronavirus-unique domain (SUD): Three-domain molecular architecture in solution and RNA binding. I: Structure of the SUD-M domain of SUD-MC
- Seq Identity
- 78.13%
- Coverage
Model-Template Alignment
Model 04
QMEANDisCo Local
QMEAN Z-Scores
Template
2k87.1.A
Non-structural protein 3 of Replicase polyprotein 1a
NMR STRUCTURE OF A PUTATIVE RNA BINDING PROTEIN (SARS1) FROM SARS CORONAVIRUS
NMR STRUCTURE OF A PUTATIVE RNA BINDING PROTEIN (SARS1) FROM SARS CORONAVIRUS
- Seq Identity
- 81.74%
- Coverage
Model-Template Alignment
Model 03
QMEANDisCo Local
QMEAN Z-Scores
Template
2kqw.1.A
Non-structural protein 3
SARS coronavirus-unique domain (SUD): Three-domain molecular architecture in solution and RNA binding. II: Structure of the SUD-C domain of SUD-MC
SARS coronavirus-unique domain (SUD): Three-domain molecular architecture in solution and RNA binding. II: Structure of the SUD-C domain of SUD-MC
- Seq Identity
- 80.22%
- Coverage
Model-Template Alignment