| UniProtKB AC (Name) | UniProtKB Section | Organism | Description | |
|---|---|---|---|---|
| P0DTD8 (NS7B_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF7b protein; Accessory protein 7b; | |
| P0DTC8 (NS8_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF8 protein; Non-structural protein 8; | |
| A0A663DJA2 (ORF10_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Putative ORF10 protein; | |
| P0DTD2 (ORF9B_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF9b protein; Accessory protein 9b; ORF-9b; Protein 9b; | |
| P0DTD3 (ORF9C_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Putative ORF9c protein; Uncharacterized protein 14; | |
| P0DTC3 (AP3A_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; Accessory protein 3a; Protein 3a; Protein U274; Protein X1; | |
| P0DTC4 (VEMP_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Envelope small membrane protein; | |
| P0DTC5 (VME1_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Membrane protein; E1 glycoprotein; Matrix glycoprotein; Membrane glycoprotein; | |
| P0DTC9 (NCAP_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; | |
| P0DTC6 (NS6_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF6 protein; Accessory protein 6; Non-structural protein 6; Protein X3; | |
| P0DTC7 (NS7A_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF7a protein; Accessory protein 7a; Protein U122; Protein X4; | |
| P0DTC2 (SPIKE_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| P0DTC1 (R1A_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Replicase polyprotein 1a; ORF1a polyprotein; Host translation inhibitor nsp1; Leader protein; Non-structural protein 1; nsp1; Non-structural protein 2; nsp2; p65 homolog; Papain-like protease nsp3; 3.4.19.12; 3.4.22.-; Non-structural protein 3; nsp3; PL2-PRO; Papain-like proteinase; PL-PRO; Non-structural protein 4; nsp4; 3C-like proteinase nsp5; 3CL-PRO; 3CLp; 3.4.22.69; Main protease; Mpro; Non-structural protein 5; nsp5; SARS coronavirus main proteinase; Non-structural protein 6; nsp6; Non-structural protein 7; nsp7; Non-structural protein 8; nsp8; RNA-capping enzyme subunit nsp9; Non-structural protein 9; nsp9; 2.7.7.50; Non-structural protein 10; nsp10; Growth factor-like peptide; GFL; Non-structural protein 11; nsp11; | |
| P0DTD1 (R1AB_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Replicase polyprotein 1ab; ORF1ab polyprotein; Host translation inhibitor nsp1; Leader protein; Non-structural protein 1; nsp1; Non-structural protein 2; nsp2; p65 homolog; Papain-like protease nsp3; 3.4.19.12; 3.4.22.-; Non-structural protein 3; nsp3; PL2-PRO; Papain-like proteinase; PL-PRO; Non-structural protein 4; nsp4; 3C-like proteinase nsp5; 3CL-PRO; 3CLp; 3.4.22.69; Main protease; Mpro; Non-structural protein 5; nsp5; SARS coronavirus main proteinase; Non-structural protein 6; nsp6; Non-structural protein 7; nsp7; Non-structural protein 8; nsp8; Viral protein genome-linked nsp9; Non-structural protein 9; nsp9; RNA-capping enzyme subunit nsp9; Non-structural protein 10; nsp10; Growth factor-like peptide; GFL; RNA-directed RNA polymerase nsp12; Pol; RdRp; 2.7.7.48; 2.7.7.50; Non-structural protein 12; nsp12; Helicase nsp13; Hel; 3.6.4.12; 3.6.4.13; Non-structural protein 13; nsp13; Guanine-N7 methyltransferase nsp14; 2.1.1.56; 3.1.13.-; Non-structural protein 14; nsp14; Proofreading exoribonuclease nsp14; ExoN; Uridylate-specific endoribonuclease nsp15; 4.6.1.-; NendoU; Non-structural protein 15; nsp15; 2'-O-methyltransferase nsp16; 2.1.1.57; Non-structural protein 16; nsp16; | |
| P0DTG1 (ORF3C_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3c protein; ORF3h protein; | |
| P0DTG0 (ORF3D_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Putative ORF3d protein; | |
| P0DTF1 (ORF3B_SARS2) | Swiss-Prot | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Putative ORF3b protein; | |
| A0A8B1JJI7 (A0A8B1JJI7_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B6RLQ1 (A0A8B6RLQ1_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; | |
| A0A8A4Y2B6 (A0A8A4Y2B6_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Envelope small membrane protein; | |
| A0A8B1JMB1 (A0A8B1JMB1_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; | |
| A0A8B1JY94 (A0A8B1JY94_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; | |
| A0A8B1JHR2 (A0A8B1JHR2_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B1K7E7 (A0A8B1K7E7_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; | |
| A0A8B1JRY1 (A0A8B1JRY1_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; | |
| A0A8B6RF95 (A0A8B6RF95_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Membrane protein; E1 glycoprotein; Matrix glycoprotein; Membrane glycoprotein; | |
| A0A8B6RKJ4 (A0A8B6RKJ4_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; | |
| A0A7U3ECQ5 (A0A7U3ECQ5_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF8 protein; | |
| A0A8B0G593 (A0A8B0G593_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8A5QK30 (A0A8A5QK30_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; | |
| A0A7G3XWA4 (A0A7G3XWA4_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A6M4BCN3 (A0A6M4BCN3_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; | |
| A0A7G6GKH1 (A0A7G6GKH1_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; | |
| A0A899K297 (A0A899K297_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B1JF89 (A0A8B1JF89_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; | |
| A0A8B1JHK5 (A0A8B1JHK5_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B1KEA4 (A0A8B1KEA4_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF8 protein; | |
| A0A8B1KVP9 (A0A8B1KVP9_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B6R8T4 (A0A8B6R8T4_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A7U3HE66 (A0A7U3HE66_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8A5PVA5 (A0A8A5PVA5_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; | |
| A0A8B1J8G0 (A0A8B1J8G0_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF8 protein; | |
| A0A8B1JXB3 (A0A8B1JXB3_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; | |
| A0A8B1K0H1 (A0A8B1K0H1_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B6RGN2 (A0A8B6RGN2_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A8B1JZ01 (A0A8B1JZ01_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF3a protein; | |
| A0A7G9PJ26 (A0A7G9PJ26_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | ORF7a protein-nanoluciferase; | |
| A0A7U3MRK2 (A0A7U3MRK2_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Spike glycoprotein; E2; Peplomer protein; Spike protein S1; Spike protein S2; Spike protein S2'; | |
| A0A6M4BBU4 (A0A6M4BBU4_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; | |
| A0A7U3EDT4 (A0A7U3EDT4_SARS2) | TrEMBL | Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) | Nucleoprotein; Nucleocapsid protein; |
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a positive-sense, single-stranded RNA coronavirus. The virus is the causative agent of coronavirus disease 2019 (COVID-19) and is contagious through human-to-human transmission.
"SARS-CoV-2", Wikipedia: The Free Encyclopedia
We modelled the full SARS-CoV-2 proteome based on the NCBI reference sequence NC_045512 which is identical to GenBank entry MN908947, and annotations from UniProt. The underlying SWISS-MODEL workspace projects are accessible below. Our colleagues at the Swiss Institute of Bioinformatics provide more resources on the SARS-CoV-2 genome on ViralZone or allow to track the current outbreak on Nextstrain. Several SARS-CoV-2 proteins reportedly interact in hetero-oligomeric complexes for which we also provide models in a dedicated hetero section. But be aware: the result of any theoretical modelling procedure is NON-EXPERIMENTAL and must be considered with care as models may contain significant errors. To this end SWISS-MODEL provides quality metrics which are described in the help section. The provided modelling projects are based on SWISS-MODEL template library version 2023-05-05 and will be updated as new templates become available.
IntAct lists interactions derived from literature curation or direct user submissions. We extracted those interactions and list the ones between SARS-CoV-2 and human host proteins with their structural coverage in a dedicated interaction page.
SARS-CoV-2 Variants of Concern were added as Annotation Projects to SWISS-MODEL Repository for Omicron variants 21K (BA.1), 21L (BA.2), 22A (BA.4), 22B (BA.5) and 22C (BA.2.12.1). Compare these variants on the spike protein, SPIKE_SARS2
Previously circulating Variants of Concern included Alpha, Beta, Delta and Gamma. Compare these variants on the spike protein, SPIKE_SARS2
Previously circulating Variants of Interest included Lambda and Mu. Compare these on the spike protein, SPIKE_SARS2
>sp|P0DTD1|R1AB_SARS2|1-7096 MESLVPGFNEKTHVQLSLPVLQVRDVLVRGFGDSVEEVLSEARQHLKDGTCGLVEVEKGV LPQLEQPYVFIKRSDARTAPHGHVMVELVAELEGIQYGRSGETLGVLVPHVGEIPVAYRK VLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQENWNTKHSSGVTRELMRELNGG AYTRYVDNNFCGPDGYPLECIKDLLARAGKASCTLSEQLDFIDTKRGVYCCREHEHEIAW YTERSEKSYELQTPFEIKLAKKFDTFNGECPNFVFPLNSIIKTIQPRVEKKKLDGFMGRI RSVYPVASPNECNQMCLSTLMKCDHCGETSWQTGDFVKATCEFCGTENLTKEGATTCGYL PQNAVVKIYCPACHNSEVGPEHSLAEYHNESGLKTILRKGGRTIAFGGCVFSYVGCHNKC AYWVPRASANIGCNHTGVVGEGSEGLNDNLLEILQKEKVNINIVGDFKLNEEIAIILASF SASTSAFVETVKGLDYKAFKQIVESCGNFKVTKGKAKKGAWNIGEQKSILSPLYAFASEA ARVVRSIFSRTLETAQNSVRVLQKAAITILDGISQYSLRLIDAMMFTSDLATNNLVVMAY ITGGVVQLTSQWLTNIFGTVYEKLKPVLDWLEEKFKEGVEFLRDGWEIVKFISTCACEIV GGQIVTCAKEIKESVQTFFKLVNKFLALCADSIIIGGAKLKALNLGETFVTHSKGLYRKC VKSREETGLLMPLKAPKEIIFLEGETLPTEVLTEEVVLKTGDLQPLEQPTSEAVEAPLVG TPVCINGLMLLEIKDTEKYCALAPNMMVTNNTFTLKGGAPTKVTFGDDTVIEVQGYKSVN ITFELDERIDKVLNEKCSAYTVELGTEVNEFACVVADAVIKTLQPVSELLTPLGIDLDEW SMATYYLFDESGEFKLASHMYCSFYPPDEDEEEGDCEEEEFEPSTQYEYGTEDDYQGKPL EFGATSAALQPEEEQEEDWLDDDSQQTVGQQDGSEDNQTTTIQTIVEVQPQLEMELTPVV QTIEVNSFSGYLKLTDNVYIKNADIVEEAKKVKPTVVVNAANVYLKHGGGVAGALNKATN NAMQVESDDYIATNGPLKVGGSCVLSGHNLAKHCLHVVGPNVNKGEDIQLLKSAYENFNQ HEVLLAPLLSAGIFGADPIHSLRVCVDTVRTNVYLAVFDKNLYDKLVSSFLEMKSEKQVE QKIAEIPKEEVKPFITESKPSVEQRKQDDKKIKACVEEVTTTLEETKFLTENLLLYIDIN GNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKALRKV PTDNYITTYPGQGLNGYTVEEAKTVLKKCKSAFYILPSIISNEKQEILGTVSWNLREMLA HAEETRKLMPVCVETKAIVSTIQRKYKGIKIQEGVVDYGARFYFYTSKTTVASLINTLND LNETLVTMPLGYVTHGLNLEEAARYMRSLKVPATVSVSSPDAVTAYNGYLTSSSKTPEEH FIETISLAGSYKDWSYSGQSTQLGIEFLKRGDKSVYYTSNPTTFHLDGEVITFDNLKTLL SLREVRTIKVFTTVDNINLHTQVVDMSMTYGQQFGPTYLDGADVTKIKPHNSHEGKTFYV LPNDDTLRVEAFEYYHTTDPSFLGRYMSALNHTKKWKYPQVNGLTSIKWADNNCYLATAL LTLQQIELKFNPPALQDAYYRARAGEAANFCALILAYCNKTVGELGDVRETMSYLFQHAN LDSCKRVLNVVCKTCGQQQTTLKGVEAVMYMGTLSYEQFKKGVQIPCTCGKQATKYLVQQ ESPFVMMSAPPAQYELKHGTFTCASEYTGNYQCGHYKHITSKETLYCIDGALLTKSSEYK GPITDVFYKENSYTTTIKPVTYKLDGVVCTEIDPKLDNYYKKDNSYFTEQPIDLVPNQPY PNASFDNFKFVCDNIKFADDLNQLTGYKKPASRELKVTFFPDLNGDVVAIDYKHYTPSFK KGAKLLHKPIVWHVNNATNKATYKPNTWCIRCLWSTKPVETSNSFDVLKSEDAQGMDNLA CEDLKPVSEEVVENPTIQKDVLECNVKTTEVVGDIILKPANNSLKITEEVGHTDLMAAYV DNSSLTIKKPNELSRVLGLKTLATHGLAAVNSVPWDTIANYAKPFLNKVVSTTTNIVTRC LNRVCTNYMPYFFTLLLQLCTFTRSTNSRIKASMPTTIAKNTVKSVGKFCLEASFNYLKS PNFSKLINIIIWFLLLSVCLGSLIYSTAALGVLMSNLGMPSYCTGYREGYLNSTNVTIAT YCTGSIPCSVCLSGLDSLDTYPSLETIQITISSFKWDLTAFGLVAEWFLAYILFTRFFYV LGLAAIMQLFFSYFAVHFISNSWLMWLIINLVQMAPISAMVRMYIFFASFYYVWKSYVHV VDGCNSSTCMMCYKRNRATRVECTTIVNGVRRSFYVYANGGKGFCKLHNWNCVNCDTFCA GSTFISDEVARDLSLQFKRPINPTDQSSYIVDSVTVKNGSIHLYFDKAGQKTYERHSLSH FVNLDNLRANNTKGSLPINVIVFDGKSKCEESSAKSASVYYSQLMCQPILLLDQALVSDV GDSAEVAVKMFDAYVNTFSSTFNVPMEKLKTLVATAEAELAKNVSLDNVLSTFISAARQG FVDSDVETKDVVECLKLSHQSDIEVTGDSCNNYMLTYNKVENMTPRDLGACIDCSARHIN AQVAKSHNIALIWNVKDFMSLSEQLRKQIRSAAKKNNLPFKLTCATTRQVVNVVTTKIAL KGGKIVNNWLKQLIKVTLVFLFVAAIFYLITPVHVMSKHTDFSSEIIGYKAIDGGVTRDI ASTDTCFANKHADFDTWFSQRGGSYTNDKACPLIAAVITREVGFVVPGLPGTILRTTNGD FLHFLPRVFSAVGNICYTPSKLIEYTDFATSACVLAAECTIFKDASGKPVPYCYDTNVLE GSVAYESLRPDTRYVLMDGSIIQFPNTYLEGSVRVVTTFDSEYCRHGTCERSEAGVCVST SGRWVLNNDYYRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDISASIVAGGIVAIVVTCL AYYFMRFRRAFGEYSHVVAFNTLLFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFYLTNDV SFLAHIQWMVMFTPLVPFWITIAYIICISTKHFYWFFSNYLKRRVVFNGVSFSTFEEAAL CTFLLNKEMYLKLRSDVLLPLTQYNRYLALYNKYKYFSGAMDTTSYREAACCHLAKALND FSNSGSDVLYQPPQTSITSAVLQSGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDVVY CPRHVICTSEDMLNPNYEDLLIRKSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPK TPKYKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCV SFCYMHHMELPTGVHAGTDLEGNFYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDR WFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMN GRTILGSALLEDEFTPFDVVRQCSGVTFQSAVKRTIKGTHHWLLLTILTSLLVLVQSTQW SLFFFLYENAFLPFAMGIIAMSAFAMMFVKHKHAFLCLFLLPSLATVAYFNMVYMPASWV MRIMTWLDMVDTSLSGFKLKDCVMYASAVVLLILMTARTVYDDGARRVWTLMNVLTLVYK VYYGNALDQAISMWALIISVTSNYSGVVTTVMFLARGIVFMCVEYCPIFFITGNTLQCIM LVYCFLGYFCTCYFGLFCLLNRYFRLTLGVYDYLVSTQEFRYMNSQGLLPPKNSIDAFKL NIKLLGVGGKPCIKVATVQSKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILL AKDTTEAFEKMVSLLSVLLSMQGAVDINKLCEEMLDNRATLQAIASEFSSLPSYAAFATA QEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSED KRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVIPDYNTY KNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNLAWPLIVTALRANSAVKLQ NNELSPVALRQMSCAAGTTQTACTDDNALAYYNTTKGGRFVLALLSDLQDLKWARFPKSD GTGTIYTELEPPCRFVTDTPKGPKVKYLYFIKGLNNLNRGMVLGSLAATVRLQAGNATEV PANSTVLSFCAFAVDAAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPEANMDQES FGGASCCLYCRCHIDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVCGMWKGYG CSCDQLREPMLQSADAQSFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNDKVAGFAKF LKTNCCRFQEKDEDDNLIDSYFVVKRHTFSNYQHEETIYNLLKDCPAVAKHDFFKFRIDG DMVPHISRQRLTKYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKKDWYDFVEN PDILRVYANLGERVRQALLKTVQFCDAMRNAGIVGVLTLDNQDLNGNWYDFGDFIQTTPG SGVPVVDSYYSLLMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERLKLFDRYFK YWDQTYHPNCVNCLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHF RELGVVHNQDVNLHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAALTNNVAFQ TVKPGNFNKDFYDFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQ LLFVVEVVDKYFDCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALF AYTKRNVIPTITQMNLKYAISAKNRARTVAGVSICSTMTNRQFHQKLLKSIAATRGATVV IGTSKFYGGWHNMLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLARKHTTCCSL SHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALL STDGNKIADKYVRNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSDDAVVCFNS TYASQGLVASIKNFKSVLYYQNNVFMSEAKCWTETDLTKGPHEFCSQHTMLVKQGDDYVY LPYPDPSRILGAGCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADVFHLYLQYI RKLHDELTGHMLDMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQAVGACVLCNSQTSLRC GACIRRPFLCCKCCYDHVISTSHKLVLSVNPYVCNAPGCDVTDVTQLYLGGMSYYCKSHK PPISFPLCANGQVFGLYKNTCVGSDNVTDFNAIATCDWTNAGDYILANTCTERLKLFAAE TLKATEETFKLSYGIATVREVLSDRELHLSWEVGKPRPPLNRNYVFTGYRVTKNSKVQIG EYTFEKGDYGDAVVYRGTTTYKLNVGDYFVLTSHTVMPLSAPTLVPQEHYVRITGLYPTL NISDEFSSNVANYQKVGMQKYSTLQGPPGTGKSHFAIGLALYYPSARIVYTACSHAAVDA LCEKALKYLPIDKCSRIIPARARVECFDKFKVNSTLEQYVFCTVNALPETTADIVVFDEI SMATNYDLSVVNARLRAKHYVYIGDPAQLPAPRTLLTKGTLEPEYFNSVCRLMKTIGPDM FLGTCRRCPAEIVDTVSALVYDNKLKAHKDKSAQCFKMFYKGVITHDVSSAINRPQIGVV REFLTRNPAWRKAVFISPYNSQNAVASKILGLPTQTVDSSQGSEYDYVIFTQTTETAHSC NVNRFNVAITRAKVGILCIMSDRDLYDKLQFTSLEIPRRNVATLQAENVTGLFKDCSKVI TGLHPTQAPTHLSVDTKFKTEGLCVDIPGIPKDMTYRRLISMMGFKMNYQVNGYPNMFIT REEAIRHVRAWIGFDVEGCHATREAVGTNLPLQLGFSTGVNLVAVPTGYVDTPNNTDFSR VSAKPPPGDQFKHLIPLMYKGLPWNVVRIKIVQMLSDTLKNLSDRVVFVLWAHGFELTSM KYFVKIGPERTCCLCDRRATCFSTASDTYACWHHSIGFDYVYNPFMIDVQQWGFTGNLQS NHDLYCQVHGNAHVASCDAIMTRCLAVHECFVKRVDWTIEYPIIGDELKINAACRKVQHM VVKAALLADKFPVLHDIGNPKAIKCVPQADVEWKFYDAQPCSDKAYKIEELFYSYATHSD KFTDGVCLFWNCNVDRYPANSIVCRFDTRVLSNLNLPGCDGGSLYVNKHAFHTPAFDKSA FVNLKQLPFFYYSDSPCESHGKQVVSDIDYVPLKSATCITRCNLGGAVCRHHANEYRLYL DAYNMMISAGFSLWVYKQFDTYNLWNTFTRLQSLENVAFNVVNKGHFDGQQGEVPVSIIN NTVYTKVDGVDVELFENKTTLPVNVAFELWAKRNIKPVPEVKILNNLGVDIAANTVIWDY KRDAPAHISTIGVCSMTDIAKKPTETICAPLTVFFDGRVDGQVDLFRNARNGVLITEGSV KGLQPSVGPKQASLNGVTLIGEAVKTQFNYYKKVDGVVQQLPETYFTQSRNLQEFKPRSQ MEIDFLELAMDEFIERYKLEGYAFEHIVYGDFSHSQLGGLHLLIGLAKRFKESPFELEDF IPMDSTVKNYFITDAQTGSSKCVCSVIDLLLDDFVEIIKSQDLSVVSKVVKVTIDYTEIS FMLWCKDGHVETFYPKLQSSQAWQPGVAMPNLYKMQRMLLEKCDLQNYGDSATLPKGIMM NVAKYTQLCQYLNTLTLAVPYNMRVIHFGAGSDKGVAPGTAVLRQWLPTGTLLVDSDLND FVSDADSTLIGDCATVHTANKWDLIISDMYDPKTKNVTKENDSKEGFFTYICGFIQQKLA LGGSVAIKITEHSWNADLYKLMGHFAWWTAFVTNVNASSSEAFLIGCNYLGKPREQIDGY VMHANYIFWRNTNPIQLSSYSLFDMSKFPLKLRGTAVMSLKEGQINDMILSLLSKGRLII RENNRVVISSDVLVNN
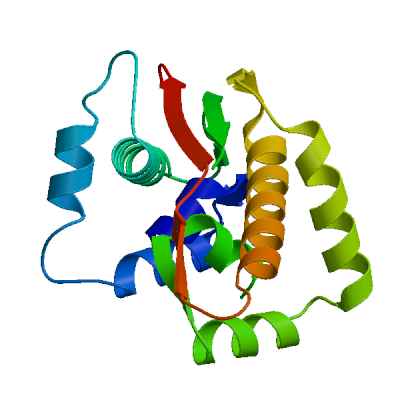
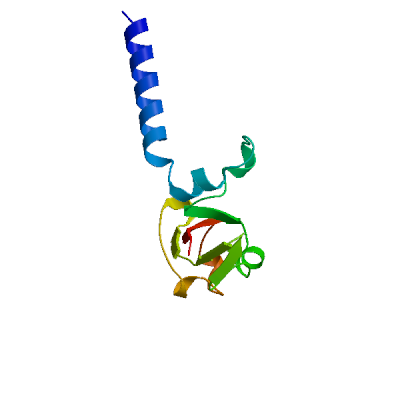
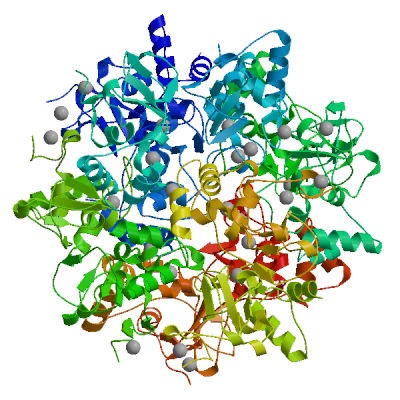
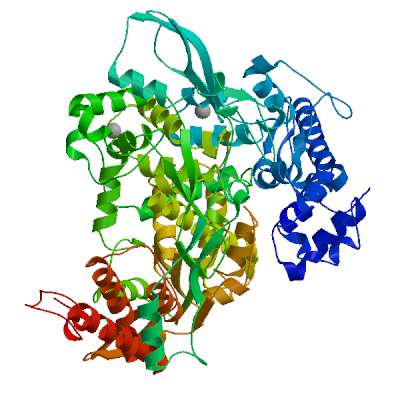
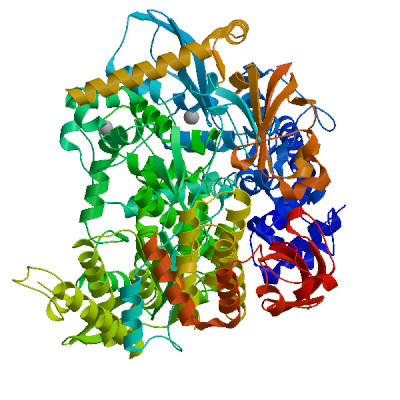
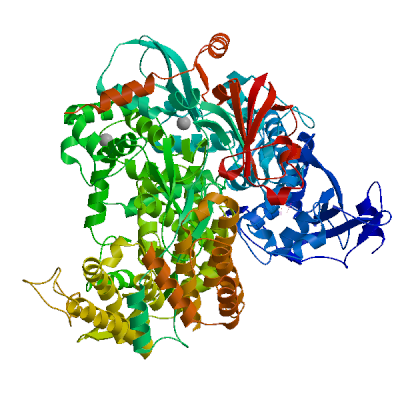
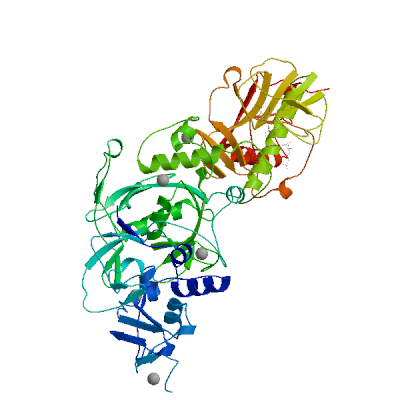
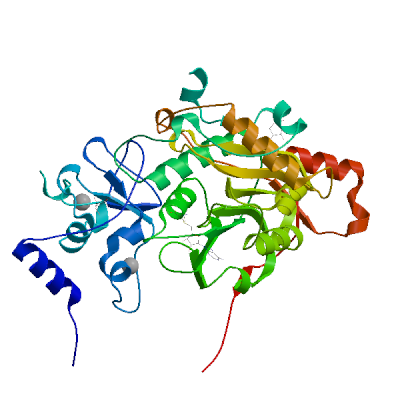
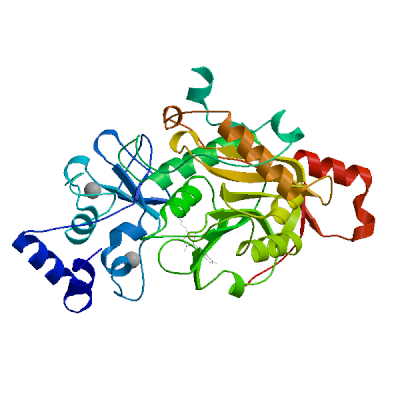
Replicase polyprotein 1ab
Multifunctional protein involved in the transcription and replication of viral RNAs. Contains the proteinases responsible for the cleavages of the polyprotein.
Partial coverage by an experimental hetero-oligomeric complex (PDB: 8eir) is available and a high quality model can be extracted but should be used with care.
We model the mature proteins resulting from the cleavage and list them below.
>sp|P0DTD1|R1AB_SARS2|1-180 MESLVPGFNEKTHVQLSLPVLQVRDVLVRGFGDSVEEVLSEARQHLKDGTCGLVEVEKGV LPQLEQPYVFIKRSDARTAPHGHVMVELVAELEGIQYGRSGETLGVLVPHVGEIPVAYRK VLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQENWNTKHSSGVTRELMRELNGG
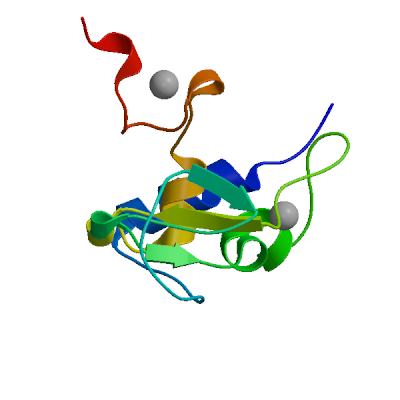
Host translation inhibitor nsp1
Inhibits host translation by associating with the open head conformation of the 40S subunit (PubMed:32680882, PubMed:32908316, PubMed:33080218, PubMed:33479166). The C-terminus binds to and obstructs ribosomal mRNA entry tunnel (PubMed:32680882, PubMed:32908316, PubMed:33080218, PubMed:33479166). Thereby inhibits antiviral response triggered by innate immunity or interferons (PubMed:32680882, PubMed:32979938, PubMed:33080218). The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation (By similarity). This inhibits the integrated stress response (ISR) in the infected cell by preventing EIF2S1/eIF2-alpha phosphorylation upstream of stress granule formation and depletes host G3BP1 (PubMed:36534661). Viral mRNAs less susceptible to nsp1-mediated inhibition of translation, because of their 5'-end leader sequence (PubMed:32908316, PubMed:33080218).
Experimental structures (PDB: 7eq4, 7ib8, 7ib9, 7iba, 7ibb, 7ibc, 7ibd, 7ibe, 7ibf, 7ibg, 7ibh, 7ibi, 7ibj, 7ibk, 7ibl, 7ibm, 7ibn, 7ibo, 7ibp, 7ibq, 7ibr, 7ibs, 7ibt, 7ibu, 7ibv, 7ibw, 7ibx, 7iby, 7ibz, 7ic0, 7ic1, 7ic2, 7ic3, 7ic4, 7ic5, 7ic6, 7ic7, 7ic8, 7ic9, 7ica, 7icb, 7icc, 7icw, 7icx, 7icy, 7icz, 7id0, 7id1, 7id2, 7id3, 7id4, 7id5, 7id6, 7id7, 7id8, 7id9, 7ida, 7idb, 7idc, 7idd, 7ide, 7idf, 7idg, 7idh, 7idi, 7idj, 7idk, 7idl, 7idm, 7idn, 7ido, 7idp, 7idq, 7idr, 7ids, 7idt, 7idu, 7idv, 7idw, 7idx, 7idy, 7idz, 7ie0, 7ie1, 7ie2, 7ie3, 7ie4, 7ie5, 7ie6, 7ie7, 7ie8, 7ie9, 7iea, 7ieb, 7iec, 7ied, 7iee, 7ief, 7ieg, 7ieh, 7iei, 7iej, 7iek, 7iel, 7iem, 7ien, 7ieo, 7iph, 7ipi, 7ipj, 7ipk, 7ipl, 7ipm, 7ipn, 7ipo, 7ipp, 7ipq, 7ipr, 7ips, 7ipt, 7ipu, 7ipv, 7ipw, 7ipx, 7ipy, 7ipz, 7iq0, 7iq1, 7iq2, 7iq3, 7iq4, 7iq5, 7iq6, 7iq7, 7iq8, 7iq9, 7iqa, 7iqb, 7iqc, 7iqd, 7iqe, 7iqf, 7iqg, 7iqh, 7iqi, 7iqj, 7iqk, 7iql, 7iqm, 7iqn, 7iqo, 7iqp, 7iqq, 7iqr, 7iqs, 7iqt, 7iqu, 7iqv, 7iqw, 7iqx, 7iqy, 7iqz, 7ir0, 7ir1, 7ir2, 7ir3, 7ir4, 7ir5, 7ir6, 7ir7, 7ir8, 7ir9, 7ira, 7irb, 7irc, 7ird, 7ire, 7irf, 7irg, 7irh, 7iri, 7irj, 7irk, 7irl, 7irm, 7irn, 7iro, 7irp, 7irq, 7irr, 7irs, 7irt, 7iru, 7irv, 7irw, 7irx, 7iry, 7irz, 7is0, 7is1, 7is2, 7is3, 7is4, 7is5, 7is6, 7is7, 7is8, 7is9, 7isa, 7isb, 7isc, 7isd, 7ise, 7isf, 7isg, 7ish, 7isi, 7isj, 7isk, 7isl, 7ism, 7isn, 7iso, 7isp, 7isq, 7isr, 7iss, 7ist, 7isu, 7isv, 7isw, 7isx, 7isy, 7isz, 7it0, 7it1, 7it2, 7it3, 7it4, 7it5, 7it6, 7it7, 7it8, 7it9, 7ita, 7itb, 7itc, 7itd, 7ite, 7itf, 7itg, 7ith, 7iti, 7k3n, 7k7p, 8a4y, 8a55, 8aou, 8ays, 8az8, 8crf, 8crk, 8crm, 8rf2, 8rf3, 8rf4, 8rf5, 8rf6, 8rf8, 8rfc, 8rfd, 8rff, 9rcz) are available. Experimental structures of hetero-oligomeric complexes (PDB: 6zlw, 6zm7, 6zme, 6zmi, 6zmo, 6zmt, 6zn5, 6zoj, 6zok, 6zon, 6zp4, 7jqb, 7jqc, 7k5i, 9npx) exist and high quality models can be extracted from them but should be used with care.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|181-818 AYTRYVDNNFCGPDGYPLECIKDLLARAGKASCTLSEQLDFIDTKRGVYCCREHEHEIAW YTERSEKSYELQTPFEIKLAKKFDTFNGECPNFVFPLNSIIKTIQPRVEKKKLDGFMGRI RSVYPVASPNECNQMCLSTLMKCDHCGETSWQTGDFVKATCEFCGTENLTKEGATTCGYL PQNAVVKIYCPACHNSEVGPEHSLAEYHNESGLKTILRKGGRTIAFGGCVFSYVGCHNKC AYWVPRASANIGCNHTGVVGEGSEGLNDNLLEILQKEKVNINIVGDFKLNEEIAIILASF SASTSAFVETVKGLDYKAFKQIVESCGNFKVTKGKAKKGAWNIGEQKSILSPLYAFASEA ARVVRSIFSRTLETAQNSVRVLQKAAITILDGISQYSLRLIDAMMFTSDLATNNLVVMAY ITGGVVQLTSQWLTNIFGTVYEKLKPVLDWLEEKFKEGVEFLRDGWEIVKFISTCACEIV GGQIVTCAKEIKESVQTFFKLVNKFLALCADSIIIGGAKLKALNLGETFVTHSKGLYRKC VKSREETGLLMPLKAPKEIIFLEGETLPTEVLTEEVVLKTGDLQPLEQPTSEAVEAPLVG TPVCINGLMLLEIKDTEKYCALAPNMMVTNNTFTLKGG
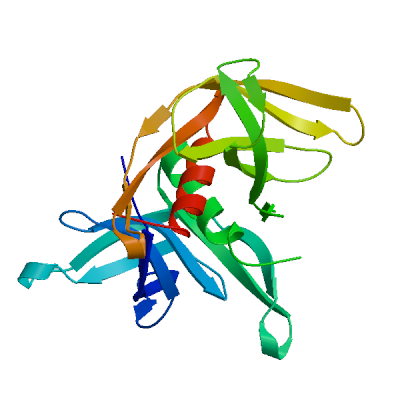
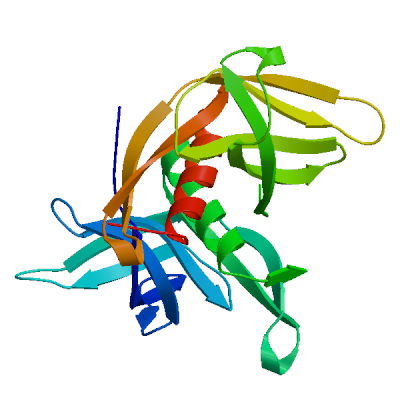
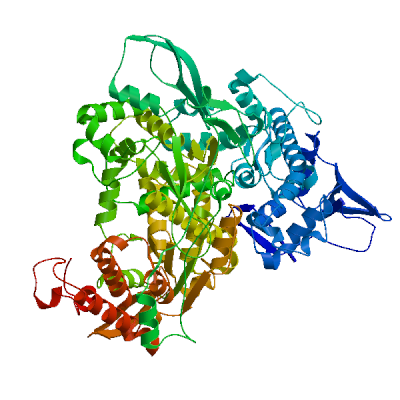
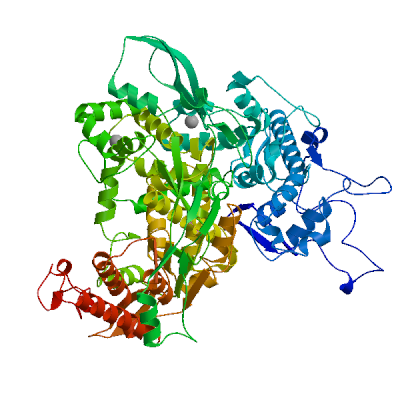
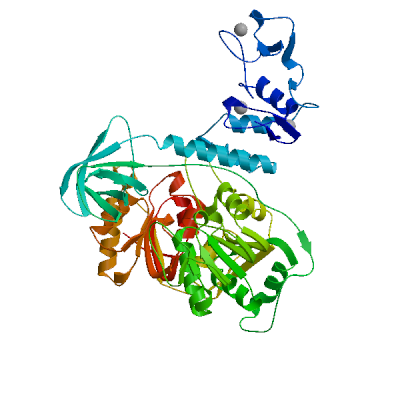
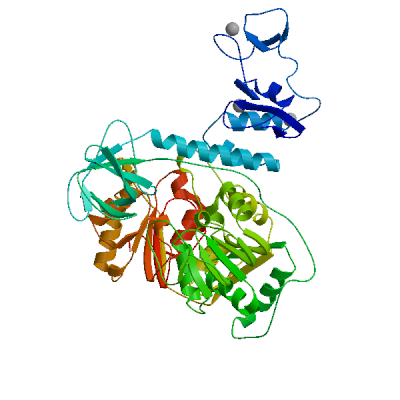
Non-structural protein 2 (nsp2)
Enhances mRNA repression of the 4EHP-GYF2 complex in the host, thereby inhibiting the antiviral response and facilitating SARS-CoV-2 replication. Possibly acts in cooperation with nsp1, which induces ribosome stalling on host mRNA, triggering mRNA repression by the host 4EHP-GYF2 complex which is enhanced by nsp2.
Experimental structures (PDB: 7exm, 7msw, 7msx) are available.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|819-2763 APTKVTFGDDTVIEVQGYKSVNITFELDERIDKVLNEKCSAYTVELGTEVNEFACVVADA VIKTLQPVSELLTPLGIDLDEWSMATYYLFDESGEFKLASHMYCSFYPPDEDEEEGDCEE EEFEPSTQYEYGTEDDYQGKPLEFGATSAALQPEEEQEEDWLDDDSQQTVGQQDGSEDNQ TTTIQTIVEVQPQLEMELTPVVQTIEVNSFSGYLKLTDNVYIKNADIVEEAKKVKPTVVV NAANVYLKHGGGVAGALNKATNNAMQVESDDYIATNGPLKVGGSCVLSGHNLAKHCLHVV GPNVNKGEDIQLLKSAYENFNQHEVLLAPLLSAGIFGADPIHSLRVCVDTVRTNVYLAVF DKNLYDKLVSSFLEMKSEKQVEQKIAEIPKEEVKPFITESKPSVEQRKQDDKKIKACVEE VTTTLEETKFLTENLLLYIDINGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTA VVIPTKKAGGTTEMLAKALRKVPTDNYITTYPGQGLNGYTVEEAKTVLKKCKSAFYILPS IISNEKQEILGTVSWNLREMLAHAEETRKLMPVCVETKAIVSTIQRKYKGIKIQEGVVDY GARFYFYTSKTTVASLINTLNDLNETLVTMPLGYVTHGLNLEEAARYMRSLKVPATVSVS SPDAVTAYNGYLTSSSKTPEEHFIETISLAGSYKDWSYSGQSTQLGIEFLKRGDKSVYYT SNPTTFHLDGEVITFDNLKTLLSLREVRTIKVFTTVDNINLHTQVVDMSMTYGQQFGPTY LDGADVTKIKPHNSHEGKTFYVLPNDDTLRVEAFEYYHTTDPSFLGRYMSALNHTKKWKY PQVNGLTSIKWADNNCYLATALLTLQQIELKFNPPALQDAYYRARAGEAANFCALILAYC NKTVGELGDVRETMSYLFQHANLDSCKRVLNVVCKTCGQQQTTLKGVEAVMYMGTLSYEQ FKKGVQIPCTCGKQATKYLVQQESPFVMMSAPPAQYELKHGTFTCASEYTGNYQCGHYKH ITSKETLYCIDGALLTKSSEYKGPITDVFYKENSYTTTIKPVTYKLDGVVCTEIDPKLDN YYKKDNSYFTEQPIDLVPNQPYPNASFDNFKFVCDNIKFADDLNQLTGYKKPASRELKVT FFPDLNGDVVAIDYKHYTPSFKKGAKLLHKPIVWHVNNATNKATYKPNTWCIRCLWSTKP VETSNSFDVLKSEDAQGMDNLACEDLKPVSEEVVENPTIQKDVLECNVKTTEVVGDIILK PANNSLKITEEVGHTDLMAAYVDNSSLTIKKPNELSRVLGLKTLATHGLAAVNSVPWDTI ANYAKPFLNKVVSTTTNIVTRCLNRVCTNYMPYFFTLLLQLCTFTRSTNSRIKASMPTTI AKNTVKSVGKFCLEASFNYLKSPNFSKLINIIIWFLLLSVCLGSLIYSTAALGVLMSNLG MPSYCTGYREGYLNSTNVTIATYCTGSIPCSVCLSGLDSLDTYPSLETIQITISSFKWDL TAFGLVAEWFLAYILFTRFFYVLGLAAIMQLFFSYFAVHFISNSWLMWLIINLVQMAPIS AMVRMYIFFASFYYVWKSYVHVVDGCNSSTCMMCYKRNRATRVECTTIVNGVRRSFYVYA NGGKGFCKLHNWNCVNCDTFCAGSTFISDEVARDLSLQFKRPINPTDQSSYIVDSVTVKN GSIHLYFDKAGQKTYERHSLSHFVNLDNLRANNTKGSLPINVIVFDGKSKCEESSAKSAS VYYSQLMCQPILLLDQALVSDVGDSAEVAVKMFDAYVNTFSSTFNVPMEKLKTLVATAEA ELAKNVSLDNVLSTFISAARQGFVDSDVETKDVVECLKLSHQSDIEVTGDSCNNYMLTYN KVENMTPRDLGACIDCSARHINAQVAKSHNIALIWNVKDFMSLSEQLRKQIRSAAKKNNL PFKLTCATTRQVVNVVTTKIALKGG
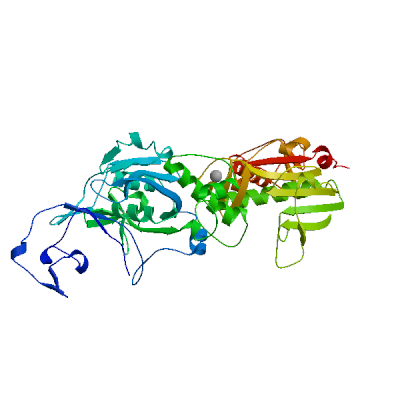
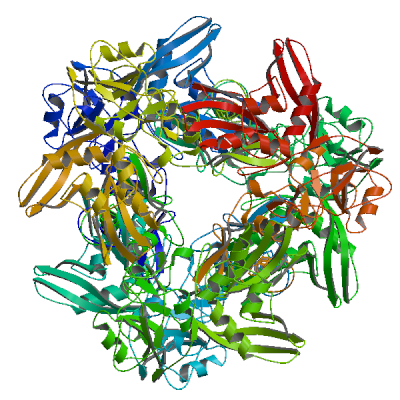
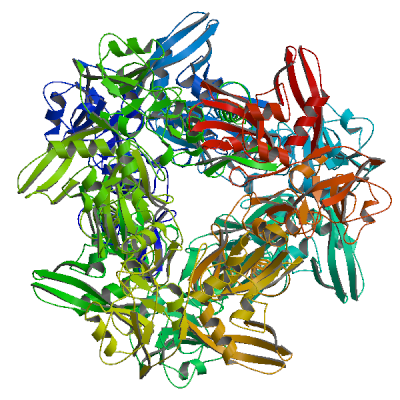
Papain-like protease nsp3
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. Participates together with nsp4 in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication (PubMed:35551511). Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3 (PubMed:32733001). Also prevents host NF-kappa-B signaling (By similarity). In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates (PubMed:32726803). Cleaves preferentially ISG15 from antiviral protein IFIH1 (MDA5), but not RIGI (PubMed:33727702). Can play a role in host ADP-ribosylation by ADP-ribose (PubMed:32578982). Plays a role in the formation and maintenance of double membrane vesicles (DMVs) replication organelles (PubMed:35551511). DMVs are formed by nsp3 and nsp4, while nsp6 zippers ER membranes and connects to lipid droplets (PubMed:35551511).
Partial coverage by experimental structures (PDB: 13mi, 13mj, 13mk, 13ml, 13mm, 13mn, 13mo, 13mp, 13mq, 13mr, 13ms, 13mt, 13mu, 13mv, 13mw, 13mx, 13my, 13mz, 13na, 13nb, 13nc, 13nd, 13ne, 13nf, 13ng, 13nh, 13ni, 13nj, 13nk, 13nl, 13nm, 13nn, 13no, 13np, 13nq, 13nr, 13ns, 13nt, 13nu, 13nv, 13nw, 13nx, 13ny, 13nz, 13oa, 13ob, 13oc, 13od, 13oe, 13of, 13og, 13oh, 13oi, 13oj, 13ok, 13ol, 13om, 13on, 13oo, 13op, 13oq, 13or, 13os, 13ot, 13ou, 13ov, 13ow, 13ox, 13oy, 13oz, 13pa, 13pb, 13pc, 13pd, 13pe, 13pf, 13pg, 13ph, 13pi, 13pj, 13pl, 13pm, 13pn, 13po, 13pp, 13pq, 13pr, 13ps, 13pt, 13pu, 13pv, 13pw, 13px, 13py, 13pz, 13qa, 13qb, 13qc, 13qd, 13qe, 13qf, 13qg, 13qh, 13qi, 13qj, 13qk, 13ql, 13qm, 13qn, 13qo, 13qp, 13qq, 13qr, 13qs, 13qt, 13qu, 13qv, 13qw, 13qx, 13qy, 13qz, 13ra, 13rb, 13rc, 13rd, 13re, 13rf, 13rg, 13rh, 13ri, 13rj, 13rk, 13rl, 13rm, 13rn, 13ro, 13rp, 13rq, 13rr, 13rs, 13rt, 13ru, 13rv, 13rw, 13rx, 13ry, 5rs7, 5rs8, 5rs9, 5rsb, 5rsc, 5rsd, 5rse, 5rsf, 5rsg, 5rsh, 5rsi, 5rsj, 5rsk, 5rsl, 5rsm, 5rsn, 5rso, 5rsp, 5rsq, 5rsr, 5rss, 5rst, 5rsu, 5rsv, 5rsw, 5rsx, 5rsy, 5rsz, 5rt0, 5rt1, 5rt2, 5rt3, 5rt4, 5rt5, 5rt6, 5rt7, 5rt8, 5rt9, 5rta, 5rtb, 5rtc, 5rtd, 5rte, 5rtf, 5rtg, 5rth, 5rti, 5rtj, 5rtk, 5rtl, 5rtm, 5rtn, 5rto, 5rtp, 5rtq, 5rtr, 5rts, 5rtt, 5rtu, 5rtv, 5rtw, 5rtx, 5rty, 5rtz, 5ru0, 5ru1, 5ru2, 5ru3, 5ru4, 5ru5, 5ru6, 5ru7, 5ru8, 5ru9, 5rua, 5ruc, 5rud, 5rue, 5ruf, 5rug, 5ruh, 5rui, 5ruj, 5ruk, 5rul, 5rum, 5run, 5ruo, 5rup, 5ruq, 5rur, 5rus, 5rut, 5ruu, 5ruv, 5ruw, 5rux, 5ruy, 5ruz, 5rv0, 5rv1, 5rv2, 5rv3, 5rv4, 5rv5, 5rv6, 5rv7, 5rv8, 5rv9, 5rva, 5rvb, 5rvc, 5rvd, 5rve, 5rvf, 5rvg, 5rvh, 5rvi, 5rvj, 5rvk, 5rvl, 5rvm, 5rvn, 5rvo, 5rvp, 5rvq, 5rvr, 5rvs, 5rvt, 5rvu, 5rvv, 5s18, 5s1a, 5s1c, 5s1e, 5s1g, 5s1i, 5s1k, 5s1m, 5s1o, 5s1q, 5s1s, 5s1u, 5s1w, 5s1y, 5s20, 5s22, 5s24, 5s26, 5s27, 5s28, 5s29, 5s2a, 5s2b, 5s2c, 5s2d, 5s2e, 5s2f, 5s2g, 5s2h, 5s2i, 5s2j, 5s2k, 5s2l, 5s2m, 5s2n, 5s2o, 5s2p, 5s2q, 5s2r, 5s2s, 5s2t, 5s2u, 5s2v, 5s2w, 5s2x, 5s2y, 5s2z, 5s30, 5s31, 5s32, 5s33, 5s34, 5s35, 5s36, 5s37, 5s38, 5s39, 5s3a, 5s3b, 5s3c, 5s3d, 5s3e, 5s3f, 5s3g, 5s3h, 5s3i, 5s3j, 5s3k, 5s3l, 5s3m, 5s3n, 5s3o, 5s3p, 5s3q, 5s3r, 5s3s, 5s3t, 5s3u, 5s3v, 5s3w, 5s3x, 5s3y, 5s3z, 5s40, 5s41, 5s42, 5s43, 5s44, 5s45, 5s46, 5s47, 5s48, 5s49, 5s4a, 5s4b, 5s4c, 5s4d, 5s4e, 5s4f, 5s4g, 5s4h, 5s4i, 5s4j, 5s4k, 5s73, 5s74, 5soi, 5soj, 5sok, 5sol, 5som, 5son, 5soo, 5sop, 5soq, 5sor, 5sos, 5sot, 5sou, 5sov, 5sow, 5sox, 5soy, 5soz, 5sp0, 5sp1, 5sp2, 5sp3, 5sp4, 5sp6, 5sp7, 5sp8, 5sp9, 5spa, 5spb, 5spc, 5spd, 5spe, 5spf, 5spg, 5sph, 5spi, 5spj, 5spk, 5spl, 5spm, 5spn, 5spo, 5spp, 5spq, 5spr, 5sps, 5spt, 5spu, 5spv, 5spw, 5spx, 5spy, 5spz, 5sq0, 5sq1, 5sq2, 5sq3, 5sq4, 5sq5, 5sq6, 5sq7, 5sq8, 5sq9, 5sqa, 5sqb, 5sqc, 5sqd, 5sqe, 5sqf, 5sqg, 5sqh, 5sqi, 5sqj, 5sqk, 5sql, 5sqm, 5sqn, 5sqo, 5sqp, 5sqq, 5sqr, 5sqs, 5sqt, 5squ, 5sqv, 5sqw, 5sqx, 5sqy, 5sqz, 5sr0, 5sr1, 5sr2, 5sr3, 5sr4, 5sr5, 5sr6, 5sr7, 5sr8, 5sr9, 5sra, 5srb, 5src, 5srd, 5sre, 5srf, 5srg, 5srh, 5sri, 5srj, 5srk, 5srl, 5srm, 5srn, 5sro, 5srp, 5srq, 5srr, 5srs, 5srt, 5sru, 5srv, 5srw, 5srx, 5sry, 5srz, 5ss0, 5ss1, 5ss2, 5ss3, 5ss4, 5ss5, 5ss6, 5ss7, 5ss8, 5ss9, 5ssa, 5ssb, 5ssc, 5ssd, 5sse, 5ssf, 5ssg, 5ssh, 5ssi, 5ssj, 5ssk, 5ssl, 5ssm, 5ssn, 5sso, 5ssp, 5ssq, 5ssr, 6vxs, 6w02, 6w6y, 6w9c, 6wcf, 6wen, 6wey, 6woj, 6wrh, 6wuu, 6wx4, 6wzu, 6xg3, 6ywk, 6ywl, 6ywm, 6z5t, 6z6i, 6z72, 7bf3, 7bf4, 7bf5, 7bf6, 7c33, 7cjd, 7cjm, 7cmd, 7cz4, 7d47, 7d6h, 7d7k, 7d7l, 7e35, 7fr0, 7fr1, 7fr2, 7fr3, 7fr4, 7fr5, 7fr6, 7fr7, 7fr8, 7fr9, 7fra, 7frb, 7frc, 7frd, 7gyy, 7gyz, 7gz0, 7gz1, 7gz2, 7gz3, 7gz4, 7gz5, 7gz6, 7gz7, 7gz8, 7gz9, 7gza, 7gzb, 7gzc, 7gzd, 7gze, 7gzf, 7gzg, 7gzh, 7gzi, 7gzj, 7gzk, 7gzl, 7gzm, 7gzn, 7gzo, 7gzq, 7gzr, 7gzs, 7gzt, 7gzu, 7gzv, 7gzw, 7gzx, 7gzy, 7gzz, 7h00, 7h01, 7h02, 7h03, 7h04, 7h05, 7h06, 7h07, 7h08, 7h09, 7h0a, 7h0b, 7h0c, 7h0d, 7h0e, 7h0f, 7h0g, 7h0h, 7h0i, 7h0j, 7h0k, 7h0l, 7h0m, 7h0n, 7h0o, 7h0p, 7h0q, 7h0r, 7h0s, 7h0t, 7h0u, 7h0v, 7h0w, 7h0x, 7h0y, 7h0z, 7h10, 7h11, 7h12, 7h13, 7h14, 7h15, 7h16, 7h17, 7h18, 7h19, 7h1a, 7h1b, 7h1c, 7h1d, 7h1e, 7h1f, 7h1g, 7hc4, 7hc5, 7hc6, 7hc7, 7hc8, 7hc9, 7hca, 7hcb, 7hcc, 7hcd, 7hce, 7hcf, 7hcg, 7hch, 7hci, 7hcj, 7hck, 7hcl, 7hcm, 7hcn, 7hco, 7hcp, 7hcq, 7hcr, 7hcs, 7hct, 7hcu, 7hcv, 7hcw, 7hcx, 7hcy, 7hcz, 7hd0, 7hd1, 7hd2, 7hd3, 7hd4, 7hd5, 7hd6, 7hd7, 7hd8, 7hd9, 7hda, 7hdb, 7hdc, 7hdd, 7hde, 7hdf, 7hdg, 7hdh, 7hdi, 7hdj, 7hdk, 7hdl, 7hdm, 7hdn, 7hdo, 7hdp, 7hdq, 7hdr, 7hds, 7hdt, 7hdu, 7hdv, 7hdw, 7hdx, 7hdy, 7hdz, 7he0, 7he1, 7he2, 7he3, 7he4, 7he5, 7he6, 7he7, 7he8, 7he9, 7hea, 7heb, 7hec, 7hed, 7hee, 7hef, 7heg, 7heh, 7hei, 7hej, 7hek, 7hel, 7hem, 7hen, 7heo, 7hep, 7heq, 7her, 7hes, 7het, 7heu, 7hev, 7hew, 7hex, 7hey, 7hez, 7hf0, 7hf1, 7hf2, 7hf3, 7hf4, 7hf5, 7hf6, 7hf7, 7hf8, 7hf9, 7hfa, 7hfb, 7hfc, 7hfd, 7hfe, 7hff, 7hfg, 7hfh, 7hfi, 7hfj, 7hfk, 7hfl, 7hfm, 7hfn, 7hfo, 7hfp, 7hfq, 7hfr, 7hfs, 7hft, 7hfu, 7hfv, 7hfw, 7hfx, 7hfy, 7hfz, 7hhs, 7hht, 7hhu, 7hhv, 7hhw, 7hhx, 7hhy, 7hhz, 7hi0, 7hi1, 7hi2, 7hi3, 7hi4, 7hi5, 7hi6, 7hi7, 7hpi, 7hpj, 7hpk, 7hpl, 7hpm, 7hpn, 7hpo, 7hpp, 7hpq, 7hpr, 7hps, 7hpt, 7hpu, 7hpv, 7hpw, 7hpx, 7hpy, 7hpz, 7hq0, 7hq1, 7hq2, 7hq3, 7hq4, 7hq5, 7hq6, 7hq7, 7hq8, 7hq9, 7hqa, 7hqb, 7hqc, 7hqd, 7hqe, 7hqf, 7hqg, 7hqh, 7hqi, 7hqj, 7hqk, 7hql, 7hqm, 7hqn, 7hqo, 7hqp, 7iiw, 7iix, 7iiy, 7iiz, 7ij0, 7ij1, 7ij2, 7ij3, 7ij4, 7ij5, 7ij6, 7ij7, 7ij8, 7ij9, 7ija, 7ijb, 7ijc, 7ijd, 7ije, 7ijf, 7ijg, 7ijh, 7iji, 7ijj, 7ijk, 7ijl, 7ijm, 7ijn, 7ijo, 7ijp, 7ijq, 7ijr, 7ijs, 7ijt, 7iju, 7ijv, 7ijw, 7ijx, 7ijy, 7ijz, 7ik0, 7ik1, 7ik2, 7ik3, 7ik4, 7ik5, 7ik6, 7ik7, 7ik8, 7jir, 7jit, 7jiv, 7jiw, 7jme, 7jn2, 7jrn, 7kag, 7kg3, 7koj, 7kok, 7kol, 7kqo, 7kqp, 7kqw, 7kr0, 7kr1, 7krx, 7kxb, 7lbr, 7lbs, 7lg7, 7lgo, 7llf, 7llz, 7los, 7m1y, 7nfv, 7nt4, 7ofs, 7oft, 7ofu, 7p2o, 7qcg, 7qch, 7qci, 7qcj, 7qck, 7qcm, 7qg7, 7rqg, 7rzc, 7sdr, 7sgu, 7sgv, 7sgw, 7sqe, 7t9w, 7thh, 7ti9, 7twf, 7twg, 7twh, 7twi, 7twj, 7twn, 7two, 7twp, 7twq, 7twr, 7tws, 7twt, 7twv, 7tww, 7twx, 7twy, 7tx0, 7tx1, 7tx3, 7tx4, 7tx5, 7tzj, 7xc3, 7xc4, 7ybg, 8azc, 8azd, 8azi, 8azl, 8azm, 8azn, 8azo, 8azp, 8c19, 8c1a, 8ers, 8eua, 8f2e, 8fwn, 8fwo, 8g62, 8gia, 8gqc, 8hbl, 8hda, 8iho, 8ilc, 8jux, 8sh6, 8sh8, 8tv6, 8tv7, 8ufm, 8uob, 8uuf, 8uug, 8uuh, 8uuu, 8uuv, 8uuw, 8uuy, 8uvm, 8vec, 8x1x, 8xab, 8xtd, 8yx2, 8yx3, 8yx4, 8yx5, 8z4w, 8zse, 9azx, 9bf7, 9bf8, 9brv, 9brw, 9brx, 9csy, 9cy0, 9cyb, 9cyc, 9cyd, 9cyk, 9d2k, 9d6b, 9d6g, 9d6h, 9d6i, 9dnu, 9dnv, 9do1, 9do3, 9do5, 9doi, 9f7p, 9f7q, 9f7r, 9f7s, 9f7t, 9f7u, 9f7y, 9gub, 9hhg, 9hhh, 9hhi, 9or4, 9puh, 9puj, 9puy, 9pv6, 9pv9, 9pvi, 9pvk, 9u7d, 9vao, 9vwy, 9z0c, 9z0d, 9z6b, 9z6c) is available. Experimental structures of hetero-oligomeric complexes (PDB: 6xa9, 6xaa, 6yva, 7pku, 7rbr, 7rbs, 7uv5, 7wzo, 8cx9, 8yax, 8yb5, 8yb7) exist and high quality models can be extracted from them but should be used with care.
High quality models are available that cover various stretches of the target sequence.
>sp|P0DTD1|R1AB_SARS2|2764-3263 KIVNNWLKQLIKVTLVFLFVAAIFYLITPVHVMSKHTDFSSEIIGYKAIDGGVTRDIAST DTCFANKHADFDTWFSQRGGSYTNDKACPLIAAVITREVGFVVPGLPGTILRTTNGDFLH FLPRVFSAVGNICYTPSKLIEYTDFATSACVLAAECTIFKDASGKPVPYCYDTNVLEGSV AYESLRPDTRYVLMDGSIIQFPNTYLEGSVRVVTTFDSEYCRHGTCERSEAGVCVSTSGR WVLNNDYYRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDISASIVAGGIVAIVVTCLAYY FMRFRRAFGEYSHVVAFNTLLFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFYLTNDVSFL AHIQWMVMFTPLVPFWITIAYIICISTKHFYWFFSNYLKRRVVFNGVSFSTFEEAALCTF LLNKEMYLKLRSDVLLPLTQYNRYLALYNKYKYFSGAMDTTSYREAACCHLAKALNDFSN SGSDVLYQPPQTSITSAVLQ
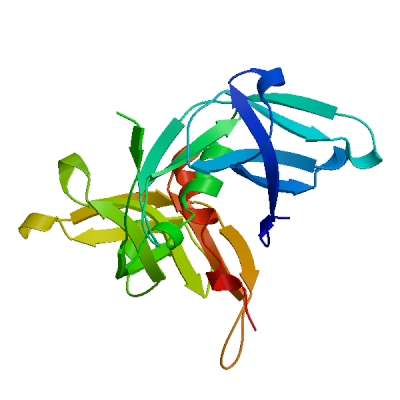
Non-structural protein 4 (nsp4)
Plays a role in the formation and maintenance of double membrane vesicles (DMVs) replication organelles (PubMed:35551511). DMVs are formed by nsp3 and nsp4, while nsp6 zippers ER membranes and connects to lipid droplets (PubMed:35551511).
Partial coverage by experimental structures (PDB: 9p0f, 9ucn) is available. Experimental structures of hetero-oligomeric complexes (PDB: 7lmc, 7mb4, 8yax, 8yb5, 8yb7) exist and high quality models can be extracted from them but should be used with care.
High quality models are available for the C-terminus which are predicted to form homo-dimers.
>sp|P0DTD1|R1AB_SARS2|3264-3569 SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDVVYCPRHVICTSEDMLNPNYEDLLIR KSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPKTPKYKFVRIQPGQTFSVLACYNG SPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMELPTGVHAGTDLEGN FYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYE PLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRTILGSALLEDEFTPFDVVRQC SGVTFQ
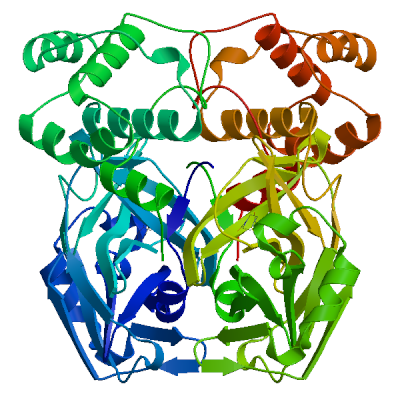
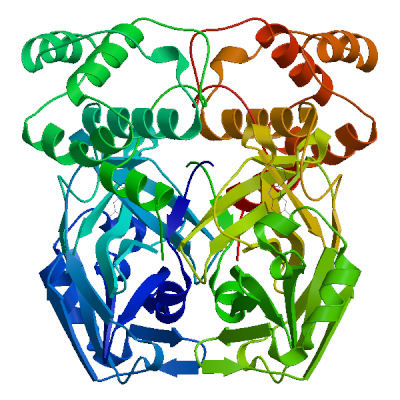
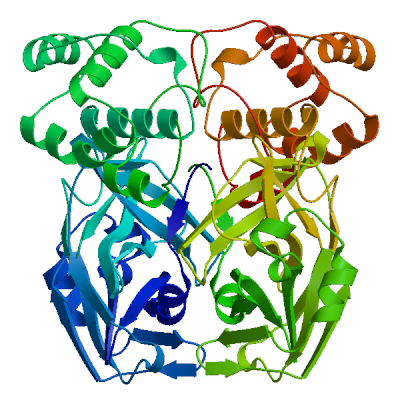
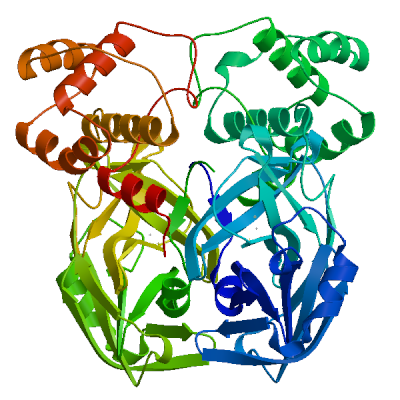
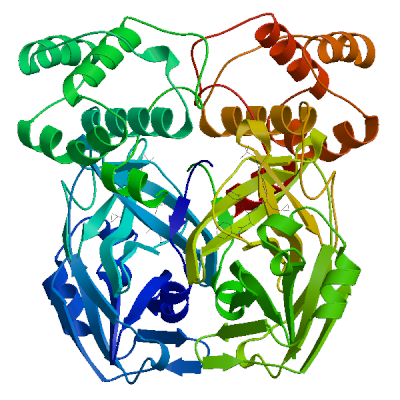
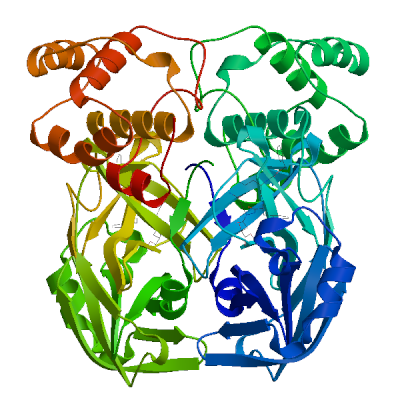
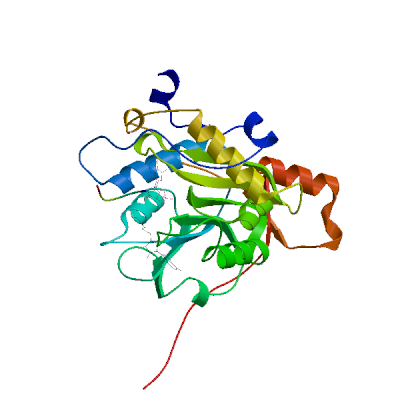
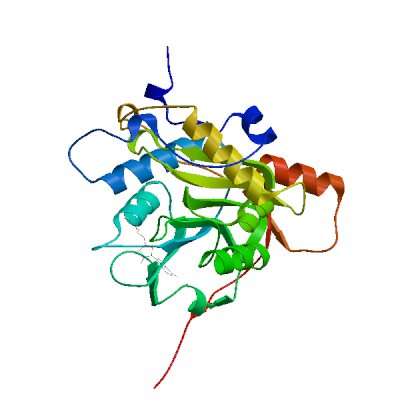
3C-like proteinase nsp5 (3CL-PRO)
Cleaves the C-terminus of replicase polyprotein at 11 sites (PubMed:32321856). Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN] (PubMed:32198291, PubMed:32272481). Cleaves and inactivates human TRMT1, preventing tRNA guanine(26)-dimethylation of tRNAs (PubMed:37073102, PubMed:38814682, PubMed:39773525). May cleave human NLRP1 in lung epithelial cells, thereby activating the NLRP1 inflammasome pathway (PubMed:35594856). May cleave human GSDMD, triggering alternative GSDME-mediated epithelial cell death upon activation of the NLRP1 inflammasome, which may enhance the release interleukins 1B, 6, 16 and 18 (PubMed:35594856). Also able to bind an ADP-ribose-1''-phosphate (ADRP) (PubMed:32198291, PubMed:32272481).
Experimental structures (PDB: 5r7y, 5r7z, 5r80, 5r81, 5r82, 5r83, 5r84, 5r8t, 5re4, 5re5, 5re6, 5re7, 5re8, 5re9, 5rea, 5reb, 5rec, 5red, 5ree, 5ref, 5reg, 5reh, 5rei, 5rej, 5rek, 5rel, 5rem, 5ren, 5reo, 5rep, 5rer, 5res, 5ret, 5reu, 5rev, 5rew, 5rex, 5rey, 5rez, 5rf0, 5rf1, 5rf2, 5rf3, 5rf4, 5rf5, 5rf6, 5rf7, 5rf8, 5rf9, 5rfa, 5rfb, 5rfc, 5rfd, 5rfe, 5rff, 5rfg, 5rfh, 5rfi, 5rfj, 5rfk, 5rfl, 5rfm, 5rfn, 5rfo, 5rfp, 5rfq, 5rfr, 5rfs, 5rft, 5rfu, 5rfv, 5rfw, 5rfx, 5rfy, 5rfz, 5rg0, 5rg1, 5rg2, 5rg3, 5rgg, 5rgh, 5rgi, 5rgj, 5rgk, 5rgl, 5rgm, 5rgn, 5rgo, 5rgp, 5rgq, 5rgr, 5rgs, 5rgt, 5rgu, 5rgv, 5rgw, 5rgx, 5rgy, 5rgz, 5rh0, 5rh1, 5rh2, 5rh3, 5rh4, 5rh5, 5rh6, 5rh7, 5rh8, 5rh9, 5rha, 5rhb, 5rhc, 5rhd, 5rhe, 5rhf, 5rl0, 5rl1, 5rl2, 5rl3, 5rl4, 5rl5, 5sml, 5smm, 5smn, 6lu7, 6lze, 6m03, 6m0k, 6m2n, 6m2q, 6w63, 6w79, 6wnp, 6wqf, 6wtj, 6wtk, 6wtm, 6wtt, 6xa4, 6xb0, 6xb1, 6xb2, 6xbg, 6xbh, 6xbi, 6xch, 6xfn, 6xhm, 6xhu, 6xkf, 6xkh, 6xmk, 6xoa, 6xqs, 6xqt, 6xqu, 6xr3, 6y2e, 6y2f, 6y2g, 6y84, 6yb7, 6ynq, 6yvf, 6z2e, 6zrt, 6zru, 7a1u, 7abu, 7adw, 7aeg, 7aeh, 7af0, 7aga, 7aha, 7ak4, 7aku, 7alh, 7ali, 7amj, 7ans, 7aol, 7ap6, 7aph, 7aqe, 7aqi, 7aqj, 7ar5, 7ar6, 7arf, 7au4, 7avd, 7awr, 7aws, 7awu, 7aww, 7ax6, 7axm, 7axo, 7ay7, 7b2j, 7b2u, 7b3e, 7b5z, 7b77, 7b83, 7baj, 7bak, 7bal, 7bb2, 7be7, 7bfb, 7bgp, 7bij, 7bqy, 7bro, 7brp, 7buy, 7c2q, 7c2y, 7c6s, 7c6u, 7c7p, 7c8b, 7c8r, 7c8t, 7c8u, 7ca8, 7cam, 7cb7, 7cbt, 7com, 7cut, 7cuu, 7cwb, 7cwc, 7cx9, 7d1m, 7d1o, 7d3i, 7d64, 7dat, 7dau, 7dav, 7ddc, 7dg6, 7dgb, 7dgf, 7dgg, 7dgh, 7dgi, 7dhj, 7djr, 7dk1, 7dpp, 7dpu, 7dpv, 7e18, 7e19, 7e5x, 7e6k, 7ein, 7en8, 7en9, 7fay, 7faz, 7gav, 7gaw, 7gax, 7gay, 7gaz, 7gb0, 7gb1, 7gb2, 7gb3, 7gb4, 7gb5, 7gb6, 7gb7, 7gb8, 7gb9, 7gba, 7gbb, 7gbc, 7gbd, 7gbe, 7gbf, 7gbg, 7gbh, 7gbi, 7gbj, 7gbk, 7gbl, 7gbm, 7gbn, 7gbo, 7gbp, 7gbq, 7gbr, 7gbs, 7gbt, 7gbu, 7gbv, 7gbw, 7gbx, 7gby, 7gbz, 7gc0, 7gc1, 7gc2, 7gc3, 7gc4, 7gc5, 7gc6, 7gc7, 7gc8, 7gc9, 7gca, 7gcb, 7gcc, 7gcd, 7gce, 7gcf, 7gcg, 7gci, 7gcj, 7gck, 7gcl, 7gcm, 7gcn, 7gco, 7gcp, 7gcq, 7gcr, 7gcs, 7gct, 7gcu, 7gcv, 7gcw, 7gcx, 7gcy, 7gcz, 7gd0, 7gd1, 7gd2, 7gd3, 7gd4, 7gd5, 7gd6, 7gd7, 7gd8, 7gd9, 7gda, 7gdb, 7gdc, 7gdd, 7gde, 7gdf, 7gdg, 7gdh, 7gdi, 7gdj, 7gdk, 7gdl, 7gdm, 7gdn, 7gdo, 7gdp, 7gdq, 7gdr, 7gds, 7gdt, 7gdu, 7gdv, 7gdw, 7gdx, 7gdy, 7gdz, 7ge0, 7ge1, 7ge2, 7ge3, 7ge4, 7ge5, 7ge6, 7ge7, 7ge8, 7ge9, 7gea, 7geb, 7gec, 7ged, 7gee, 7gef, 7geg, 7geh, 7gei, 7gej, 7gek, 7gel, 7gem, 7gen, 7geo, 7geq, 7ger, 7ges, 7get, 7geu, 7gev, 7gew, 7gex, 7gey, 7gez, 7gf0, 7gf1, 7gf2, 7gf3, 7gf4, 7gf5, 7gf6, 7gf7, 7gf8, 7gf9, 7gfa, 7gfb, 7gfc, 7gfd, 7gfe, 7gff, 7gfg, 7gfh, 7gfi, 7gfj, 7gfk, 7gfl, 7gfm, 7gfn, 7gfo, 7gfp, 7gfq, 7gfr, 7gfs, 7gft, 7gfu, 7gfv, 7gfw, 7gfx, 7gfy, 7gfz, 7gg0, 7gg1, 7gg2, 7gg3, 7gg4, 7gg5, 7gg6, 7gg7, 7gg8, 7gg9, 7gga, 7ggb, 7ggc, 7ggd, 7gge, 7ggf, 7ggg, 7ggh, 7ggi, 7ggj, 7ggk, 7ggl, 7ggm, 7ggn, 7ggo, 7ggp, 7ggq, 7ggr, 7ggs, 7ggt, 7ggu, 7ggv, 7ggw, 7ggx, 7ggy, 7ggz, 7gh0, 7gh1, 7gh2, 7gh3, 7gh4, 7gh5, 7gh6, 7gh7, 7gh8, 7gh9, 7gha, 7ghb, 7ghc, 7ghd, 7ghe, 7ghf, 7ghg, 7ghh, 7ghi, 7ghj, 7ghk, 7ghl, 7ghm, 7ghn, 7gho, 7ghp, 7ghq, 7ghr, 7ghs, 7ght, 7ghu, 7ghv, 7ghw, 7ghx, 7ghy, 7ghz, 7gi0, 7gi1, 7gi2, 7gi3, 7gi4, 7gi5, 7gi6, 7gi7, 7gi8, 7gi9, 7gia, 7gib, 7gic, 7gid, 7gie, 7gif, 7gig, 7gih, 7gii, 7gij, 7gik, 7gil, 7gim, 7gin, 7gio, 7gip, 7giq, 7gir, 7gis, 7git, 7giu, 7giv, 7giw, 7gix, 7giy, 7giz, 7gj0, 7gj1, 7gj2, 7gj3, 7gj4, 7gj5, 7gj6, 7gj7, 7gj8, 7gj9, 7gja, 7gjb, 7gjc, 7gjd, 7gje, 7gjf, 7gjg, 7gjh, 7gji, 7gjj, 7gjk, 7gjl, 7gjm, 7gjn, 7gjo, 7gjp, 7gjq, 7gjr, 7gjs, 7gjt, 7gju, 7gjv, 7gjw, 7gjx, 7gjy, 7gjz, 7gk0, 7gk1, 7gk2, 7gk3, 7gk4, 7gk5, 7gk6, 7gk7, 7gk8, 7gk9, 7gka, 7gkb, 7gkc, 7gkd, 7gke, 7gkf, 7gkg, 7gkh, 7gki, 7gkj, 7gkk, 7gkl, 7gkm, 7gkn, 7gko, 7gkp, 7gkq, 7gkr, 7gks, 7gkt, 7gku, 7gkv, 7gkw, 7gkx, 7gky, 7gkz, 7gl0, 7gl1, 7gl2, 7gl3, 7gl4, 7gl5, 7gl6, 7gl7, 7gl8, 7gl9, 7gla, 7glb, 7glc, 7gld, 7gle, 7glf, 7glg, 7glh, 7gli, 7glj, 7glk, 7gll, 7glm, 7gln, 7glo, 7glp, 7glq, 7glr, 7gls, 7glt, 7glu, 7glv, 7glw, 7glx, 7gly, 7glz, 7gm0, 7gm1, 7gm2, 7gm3, 7gm4, 7gm5, 7gm6, 7gm7, 7gm8, 7gm9, 7gma, 7gmb, 7gmc, 7gmd, 7gme, 7gmf, 7gmg, 7gmh, 7gmi, 7gmj, 7gmk, 7gml, 7gmm, 7gmn, 7gmo, 7gmp, 7gmq, 7gmr, 7gms, 7gmt, 7gmu, 7gmv, 7gmw, 7gmx, 7gmy, 7gmz, 7gn0, 7gn1, 7gn2, 7gn3, 7gn4, 7gn5, 7gn6, 7gn7, 7gn8, 7gn9, 7gna, 7gnb, 7gnc, 7gnd, 7gne, 7gnf, 7gng, 7gnh, 7gni, 7gnj, 7gnk, 7gnl, 7gnm, 7gnn, 7gno, 7gnp, 7gnq, 7gnr, 7gns, 7gnt, 7gnu, 7gre, 7grf, 7grg, 7grh, 7gri, 7grj, 7grk, 7grl, 7grm, 7grn, 7gro, 7grp, 7grq, 7grr, 7grs, 7grt, 7gru, 7grv, 7grw, 7grx, 7gry, 7grz, 7gs0, 7gs1, 7gs2, 7gs3, 7gs4, 7gs5, 7gs6, 7huc, 7hud, 7hue, 7i13, 7i14, 7i15, 7i16, 7i17, 7i18, 7i19, 7i1a, 7i1c, 7i1d, 7i1e, 7i1f, 7i1g, 7i1h, 7i1i, 7i1j, 7jfq, 7jkv, 7joy, 7jp0, 7jp1, 7jpy, 7jpz, 7jq0, 7jq1, 7jq2, 7jq3, 7jq4, 7jq5, 7jr3, 7jr4, 7jst, 7jsu, 7jt0, 7jt7, 7ju7, 7jun, 7jvz, 7jw8, 7jyc, 7k0e, 7k0f, 7k3t, 7k40, 7k6d, 7k6e, 7kfi, 7khp, 7kph, 7kvl, 7kvr, 7kx5, 7kyu, 7l0d, 7l10, 7l11, 7l12, 7l13, 7l14, 7l5d, 7l8i, 7l8j, 7lb7, 7lbn, 7lco, 7lcr, 7lcs, 7lct, 7ldl, 7ldx, 7lfe, 7lfp, 7lkd, 7lke, 7lkr, 7lks, 7lkt, 7lku, 7lkv, 7lkw, 7lkx, 7lmd, 7lme, 7lmf, 7ltj, 7ltn, 7lyh, 7lyi, 7lzt, 7lzu, 7lzv, 7lzw, 7lzx, 7lzy, 7lzz, 7m00, 7m01, 7m02, 7m03, 7m04, 7m2p, 7m8m, 7m8n, 7m8o, 7m8p, 7m8x, 7m8y, 7m8z, 7m90, 7m91, 7mat, 7mau, 7mav, 7maw, 7max, 7maz, 7mb0, 7mb1, 7mb2, 7mb3, 7mbg, 7mbi, 7mhf, 7mhg, 7mhh, 7mhi, 7mhj, 7mhk, 7mhl, 7mhm, 7mhn, 7mho, 7mhp, 7mhq, 7mlf, 7mlg, 7mng, 7mpb, 7mrr, 7n44, 7n5z, 7n8c, 7nbr, 7nbs, 7nbt, 7nby, 7neo, 7nev, 7nf5, 7ng3, 7ng6, 7nij, 7nt1, 7nt2, 7nt3, 7ntq, 7nts, 7ntt, 7ntv, 7ntw, 7nuk, 7nw2, 7nwx, 7nxh, 7o46, 7p2g, 7p35, 7p51, 7pfl, 7pfm, 7phz, 7pxz, 7pzq, 7q5e, 7q5f, 7qbb, 7qka, 7ql8, 7qt5, 7qt6, 7qt7, 7qt8, 7r7h, 7rbz, 7rc0, 7rfr, 7rfs, 7rfu, 7rfw, 7rls, 7rm2, 7rmb, 7rme, 7rmt, 7rmz, 7rn0, 7rn1, 7rn4, 7rnh, 7rnk, 7rnw, 7rvm, 7rvn, 7rvo, 7rvp, 7rvq, 7rvr, 7rvs, 7rvt, 7rvu, 7rvv, 7rvw, 7rvx, 7rvy, 7rvz, 7rw0, 7rw1, 7s3k, 7s3s, 7s4b, 7s6w, 7s6x, 7s6y, 7s6z, 7s70, 7s71, 7s72, 7s73, 7s74, 7s75, 7s82, 7sd9, 7sda, 7sdc, 7set, 7sf1, 7sf3, 7sfb, 7sfh, 7sfi, 7sgh, 7sh7, 7sh8, 7sh9, 7shb, 7si9, 7t2t, 7t2v, 7t42, 7t43, 7t44, 7t45, 7t46, 7t48, 7t49, 7t4a, 7t4b, 7tdu, 7te0, 7teh, 7tek, 7tel, 7tfr, 7tgr, 7tia, 7tiu, 7tiv, 7tiw, 7tix, 7tiy, 7tiz, 7tj0, 7tll, 7tob, 7tq2, 7tq3, 7tq4, 7tq5, 7tq6, 7tuu, 7tvx, 7u28, 7u29, 7u92, 7uj9, 7ujg, 7uju, 7ukk, 7ur9, 7urb, 7us4, 7uu6, 7uu7, 7uu8, 7uu9, 7uua, 7uub, 7uuc, 7uud, 7uue, 7uug, 7uup, 7v1t, 7v7m, 7vah, 7vfa, 7vfb, 7vh8, 7vic, 7vjw, 7vjx, 7vjy, 7vjz, 7vk0, 7vk1, 7vk2, 7vk3, 7vk4, 7vk5, 7vk6, 7vk7, 7vk8, 7vlp, 7vlq, 7vth, 7vu6, 7vvp, 7vvt, 7w9g, 7whc, 7wo1, 7wo2, 7wo3, 7wof, 7woh, 7wq8, 7wq9, 7wqa, 7wqb, 7wqk, 7wym, 7wyp, 7x6j, 7x6k, 7xar, 7xb3, 7xb4, 7xq6, 7xq7, 7xrs, 7z0p, 7z2k, 7z3u, 7z4s, 7z59, 7zb6, 7zb7, 7zb8, 7zqv, 7zv5, 7zv7, 7zv8, 8a4q, 8a4t, 8acd, 8acl, 8aeb, 8aj1, 8b0s, 8b0t, 8b2t, 8bfo, 8bfq, 8bga, 8bgd, 8bs1, 8bs2, 8c9l, 8c9o, 8c9p, 8c9q, 8c9u, 8ca6, 8ca8, 8cac, 8cae, 8caj, 8cdc, 8cyu, 8cyz, 8cz4, 8cz7, 8czw, 8czx, 8d4j, 8d4k, 8d4l, 8d4m, 8d4n, 8d4p, 8dcz, 8dd1, 8dd6, 8dd9, 8ddi, 8ddm, 8dfe, 8dfn, 8dgb, 8di3, 8dib, 8dic, 8did, 8die, 8dif, 8dig, 8dih, 8dii, 8djj, 8dk8, 8dkh, 8dkj, 8dkk, 8dkl, 8dkz, 8dl9, 8dlb, 8dmd, 8dmn, 8dox, 8doy, 8dpr, 8drr, 8drs, 8drt, 8dru, 8drv, 8drw, 8drx, 8dry, 8drz, 8ds0, 8ds1, 8ds2, 8dsu, 8dt9, 8dz0, 8dz1, 8dz2, 8dz6, 8dz9, 8dza, 8dzb, 8dzc, 8e1y, 8e25, 8e26, 8e4j, 8e4r, 8e4w, 8e5c, 8e5x, 8e5z, 8e61, 8e63, 8e64, 8e65, 8e68, 8e69, 8e6a, 8ehj, 8ehk, 8ehl, 8ehm, 8ej7, 8ej9, 8eke, 8eoy, 8ey2, 8eyj, 8ezv, 8ezz, 8f02, 8f2c, 8f2d, 8f44, 8f45, 8f46, 8fig, 8fiv, 8fiw, 8ftc, 8ftl, 8fy6, 8fy7, 8gfk, 8gfn, 8gfo, 8gfr, 8gfu, 8gqt, 8gtv, 8gtw, 8gvd, 8gvy, 8gw4, 8gwj, 8gws, 8gxg, 8gxh, 8gxi, 8gzb, 8h3g, 8h3k, 8h3l, 8h4y, 8h51, 8h57, 8h5f, 8h5p, 8h6i, 8h6n, 8h7w, 8h82, 8hbk, 8hef, 8hht, 8hhu, 8hi9, 8hol, 8hom, 8hoz, 8hqf, 8hqg, 8hqh, 8hqi, 8hqj, 8htv, 8hur, 8huv, 8huw, 8hux, 8hvk, 8hvl, 8hvm, 8hvn, 8hvo, 8hvu, 8hvv, 8hvw, 8hvx, 8hvy, 8hvz, 8hzr, 8i30, 8i4s, 8ifp, 8ifq, 8ifr, 8ifs, 8ift, 8ig4, 8ig7, 8ig8, 8ig9, 8iga, 8igb, 8ign, 8igo, 8igx, 8igy, 8inq, 8int, 8inu, 8inw, 8inx, 8iny, 8j32, 8j35, 8j36, 8j37, 8j38, 8j39, 8j3a, 8j3b, 8jcj, 8jck, 8jcl, 8jcm, 8jcn, 8jco, 8jop, 8jpq, 8k67, 8k68, 8k6a, 8k6b, 8k6c, 8k6d, 8okb, 8okc, 8okk, 8okl, 8okm, 8okn, 8p54, 8p55, 8p56, 8p57, 8p58, 8p5a, 8p5b, 8p5c, 8p86, 8p87, 8ph4, 8r0v, 8r11, 8r12, 8r14, 8r16, 8r19, 8r1q, 8r24, 8r26, 8ri4, 8rjz, 8s9z, 8sg6, 8sk4, 8skh, 8spj, 8sty, 8stz, 8sxo, 8sxr, 8t7y, 8tbe, 8tpb, 8tpc, 8tpd, 8tpg, 8tph, 8tpi, 8tqh, 8tqj, 8tql, 8tqt, 8tqu, 8ty3, 8ty4, 8ty5, 8tyk, 8u25, 8u3o, 8u40, 8u4y, 8u9h, 8u9k, 8u9m, 8u9n, 8u9t, 8u9u, 8u9v, 8u9w, 8uab, 8udf, 8udj, 8udm, 8udo, 8udp, 8udq, 8udw, 8udx, 8udy, 8ue0, 8uea, 8ueb, 8uef, 8ueg, 8ueh, 8uei, 8uh5, 8uh8, 8uh9, 8uho, 8uia, 8uif, 8ups, 8upv, 8upw, 8ur9, 8ute, 8v4u, 8v7t, 8v7w, 8v8e, 8v8g, 8vd7, 8vdj, 8vqx, 8vsg, 8w1t, 8w1u, 8ws3, 8wsh, 8wsi, 8wsk, 8wti, 8wts, 8wur, 8wz0, 8wzp, 8wzq, 8xwr, 8xwt, 8y42, 8y44, 8y4d, 8y4g, 8y4h, 8y7t, 8y7u, 8ya5, 8ykj, 8ykm, 8ykn, 8yko, 8ykp, 8ykq, 8yls, 8yrh, 8ysa, 8ywy, 8ywz, 8z1h, 8z46, 8zbp, 8zq8, 8zt9, 8zub, 9arq, 9ars, 9art, 9asv, 9asw, 9asy, 9asz, 9at0, 9at1, 9at3, 9at4, 9at5, 9at6, 9at7, 9auj, 9auk, 9aul, 9aum, 9aun, 9auo, 9avq, 9bbp, 9bbq, 9bbr, 9bbs, 9bbt, 9bbu, 9bbv, 9bbw, 9bbx, 9bby, 9bbz, 9bc0, 9bc1, 9bnt, 9bnu, 9bnv, 9bnw, 9bnx, 9bny, 9bnz, 9bo0, 9bo1, 9bo2, 9bo3, 9bo4, 9bo5, 9bo6, 9bo7, 9bo8, 9bo9, 9boa, 9bob, 9boc, 9bod, 9boe, 9bpf, 9bqf, 9bqg, 9bql, 9bqm, 9bqn, 9bqo, 9bqp, 9bqq, 9bqt, 9bqy, 9bqz, 9br0, 9br1, 9bs7, 9bs8, 9bsa, 9bse, 9bsf, 9bsg, 9bsi, 9bso, 9bsp, 9bsq, 9bsr, 9bst, 9bte, 9btf, 9btk, 9btr, 9btt, 9bvw, 9bvx, 9bvz, 9c80, 9c8q, 9cdk, 9cdl, 9cdm, 9cec, 9ced, 9cek, 9cf9, 9cfb, 9cjo, 9cjp, 9cjq, 9cjr, 9cjs, 9cjt, 9cju, 9cjv, 9cmj, 9cmn, 9cms, 9cmu, 9d08, 9ddf, 9ddg, 9dtz, 9du2, 9du3, 9du4, 9e7b, 9e7s, 9eei, 9eet, 9eev, 9el4, 9elv, 9eo6, 9eor, 9eox, 9epl, 9epm, 9ex8, 9f2x, 9f39, 9f3a, 9fhq, 9fq9, 9fqa, 9fx6, 9fx7, 9g0h, 9g0i, 9gf7, 9ghn, 9gho, 9gi6, 9gij, 9gil, 9glv, 9gmq, 9gv2, 9h0f, 9h4b, 9haj, 9hak, 9hbq, 9hc1, 9hd8, 9hdc, 9hdj, 9hdn, 9hfx, 9hfy, 9hjh, 9ik2, 9inl, 9ir9, 9izb, 9j19, 9j8t, 9j8u, 9kgj, 9kgn, 9kgq, 9kgr, 9kgs, 9kr5, 9ksh, 9ksi, 9ksj, 9ksk, 9l13, 9lll, 9luf, 9lug, 9lvr, 9lvt, 9lvv, 9m2v, 9m6q, 9m6r, 9m6s, 9m6t, 9m6u, 9m6v, 9m9n, 9m9r, 9ma3, 9ma6, 9mdq, 9mei, 9mlj, 9mvm, 9mvo, 9mvp, 9mvq, 9n3m, 9n5q, 9n6f, 9n6j, 9n6l, 9n6m, 9n6n, 9n6p, 9n6r, 9n99, 9n9b, 9nng, 9nnw, 9nsk, 9nsl, 9nu6, 9nwa, 9nwc, 9o6d, 9o6e, 9o6f, 9o6p, 9o6q, 9o74, 9ock, 9oix, 9oiz, 9ojg, 9ojt, 9opm, 9opn, 9pa9, 9pbc, 9pfh, 9pfi, 9pjg, 9pkr, 9pys, 9pyt, 9q7s, 9rhs, 9rht, 9rhx, 9ri0, 9ri1, 9ri3, 9ri4, 9ri5, 9ri8, 9rid, 9rix, 9riy, 9riz, 9rj0, 9rj3, 9rj5, 9rj7, 9rj8, 9rjf, 9rjr, 9sdm, 9u96, 9uoq, 9vs1, 9vuw, 9whe, 9xym, 9xyx, 9xyz, 9xz6, 9z74, 9znl, 9zo3) are available. Experimental structures of hetero-oligomeric complexes (PDB: 7dvp, 7dvw, 7dvx, 7dvy, 7dw0, 7dw6, 7lmc, 7mb4, 7mb5, 7mb6, 7mb7, 7mb8, 7mb9, 7mgr, 7mgs, 7n6n, 7n89, 7t2u, 7t70, 7t8m, 7t8r, 7t9y, 7ta4, 7ta7, 7tb2, 7tbt, 7tc4, 7x6y, 8ddl, 8eir, 8h7k, 8wsj, 8xwr, 8zuc, 9dw6, 9exu, 9eya, 9ez4, 9ez6, 9jj7, 9lgq) exist and high quality models can be extracted from them but should be used with care.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|3570-3859 SAVKRTIKGTHHWLLLTILTSLLVLVQSTQWSLFFFLYENAFLPFAMGIIAMSAFAMMFV KHKHAFLCLFLLPSLATVAYFNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAV VLLILMTARTVYDDGARRVWTLMNVLTLVYKVYYGNALDQAISMWALIISVTSNYSGVVT TVMFLARGIVFMCVEYCPIFFITGNTLQCIMLVYCFLGYFCTCYFGLFCLLNRYFRLTLG VYDYLVSTQEFRYMNSQGLLPPKNSIDAFKLNIKLLGVGGKPCIKVATVQ
Non-structural protein 6 (nsp6)
Plays a role in the formation and maintenance of double membrane vesicles (DMVs) replication organelles (PubMed:35551511). DMVs are formed by nsp3 and nsp4, while nsp6 zippers ER membranes and connects to lipid droplets (PubMed:35551511). LDs are consumed during DMV formation (PubMed:35551511). Binds to host TBK1 without affecting TBK1 phosphorylation; the interaction with TBK1 decreases IRF3 phosphorylation, which leads to reduced IFN-beta production (PubMed:32979938).
Only remote homologues were identified as potential template structures, no models have been built.
See here for template search results.
>sp|P0DTD1|R1AB_SARS2|3860-3942 SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLL SMQGAVDINKLCEEMLDNRATLQ
Non-structural protein 7 (nsp7)
Plays a role in viral RNA synthesis (PubMed:32277040, PubMed:32358203, PubMed:32438371, PubMed:32526208). Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers (By similarity).
An experimental structure (PDB: 7lhq) is available. Experimental structures of hetero-oligomeric complexes (PDB: 6m5i, 6m71, 6wiq, 6wqd, 6wtc, 6xez, 6xip, 6xqb, 6yhu, 6yyt, 7aap, 7b3b, 7b3c, 7b3d, 7btf, 7bv1, 7bv2, 7bw4, 7bzf, 7c2k, 7ctt, 7cxm, 7cxn, 7cyq, 7d4f, 7dcd, 7dfg, 7dfh, 7doi, 7dok, 7dte, 7egq, 7eiz, 7jlt, 7krn, 7kro, 7krp, 7l1f, 7lg3, 7mb7, 7oyg, 7ozu, 7ozv, 7rdx, 7rdy, 7rdz, 7re0, 7re1, 7re2, 7re3, 7thm, 7uo4, 7uo7, 7uo9, 7uob, 7uoe, 8gw1, 8gwb, 8gwe, 8gwf, 8gwg, 8gwi, 8gwk, 8gwm, 8gwn, 8gwo, 8gy6, 8sq9, 8sqj, 8sqk, 8xch, 9blf, 9cgv, 9i81, 9ikz, 9imk, 9imm, 9l09, 9pyw, 9pyz, 9pz0, 9sao, 9sap, 9saq, 9sar, 9uht) exist and high quality models can be extracted from them but should be used with care.
>sp|P0DTD1|R1AB_SARS2|3943-4140 AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLE KMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNII PLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNL AWPLIVTALRANSAVKLQ
Non-structural protein 8 (nsp8)
Plays a role in viral RNA synthesis (PubMed:32277040, PubMed:32358203, PubMed:32438371, PubMed:32526208). Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers (By similarity). Interacts with ribosome signal recognition particle RNA (SRP) (PubMed:33080218). Together with NSP9, suppress protein integration into the cell membrane, thereby disrupting host immune defenses (PubMed:33080218).
Partial coverage by an experimental structure (PDB: 7ywr) is available. Experimental structures of hetero-oligomeric complexes (PDB: 6m5i, 6m71, 6wiq, 6wqd, 6wtc, 6xez, 6xip, 6xqb, 6yhu, 6yyt, 7aap, 7b3b, 7b3c, 7b3d, 7btf, 7bv1, 7bv2, 7bw4, 7bzf, 7c2k, 7ctt, 7cxm, 7cxn, 7cyq, 7d4f, 7dcd, 7dfg, 7dfh, 7doi, 7dok, 7dte, 7ed5, 7egq, 7eiz, 7jlt, 7krn, 7kro, 7krp, 7l1f, 7lg2, 7mb8, 7oyg, 7ozu, 7ozv, 7rdx, 7rdy, 7rdz, 7re0, 7re1, 7re2, 7re3, 7thm, 7uo4, 7uo7, 7uo9, 7uob, 7uoe, 7x70, 8gw1, 8gwb, 8gwe, 8gwf, 8gwg, 8gwi, 8gwk, 8gwm, 8gwn, 8gwo, 8gy6, 8sq9, 8sqj, 8sqk, 8xch, 9blf, 9cgv, 9i81, 9ikz, 9imk, 9imm, 9l09, 9pyw, 9pyz, 9pz0, 9sao, 9sap, 9saq, 9sar, 9uht) exist and high quality models can be extracted from them but should be used with care.
>sp|P0DTD1|R1AB_SARS2|4141-4253 NNELSPVALRQMSCAAGTTQTACTDDNALAYYNTTKGGRFVLALLSDLQDLKWARFPKSD GTGTIYTELEPPCRFVTDTPKGPKVKYLYFIKGLNNLNRGMVLGSLAATVRLQ
Viral protein genome-linked nsp9
Forms a primer, NSP9-pU, which is utilized by the polymerase for the initiation of RNA chains (PubMed:37794589). Interacts with ribosome signal recognition particle RNA (SRP) (PubMed:33080218). Together with NSP8, suppress protein integration into the cell membrane, thereby disrupting host immune defenses (PubMed:33080218).
Experimental structures (PDB: 6w4b, 6w9q, 6wc1, 6wxd, 7bwq, 7kri, 7n3k, 8dqu, 8wke) are available. Experimental structures of hetero-oligomeric complexes (PDB: 7cyq, 7egq, 7eiz, 7thm, 8gw1, 8gwb, 8gwe, 8gwf, 8gwg, 8gwi, 8gwk, 8gwm, 8gwn, 8gwo, 8sq9, 8sqj, 8sqk, 9dj8, 9ikz, 9imk, 9imm, 9uht) exist and high quality models can be extracted from them but should be used with care.
High quality models are available which are predicted to form homo-dimers.
>sp|P0DTD1|R1AB_SARS2|4254-4392 AGNATEVPANSTVLSFCAFAVDAAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPE ANMDQESFGGASCCLYCRCHIDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVC GMWKGYGCSCDQLREPMLQ
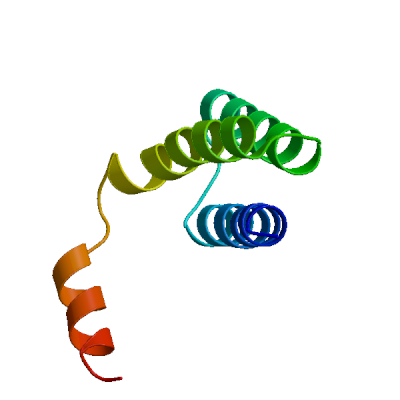
Non-structural protein 10 (nsp10)
Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease (By similarity) and nsp16 2'-O-methyltransferase activities (PubMed:35944563). Therefore plays an essential role in viral mRNAs cap methylation.
Experimental structures (PDB: 6zct, 6zpe, 7orr, 7oru, 7orv, 7orw, 8bzn) are available. Experimental structures of hetero-oligomeric complexes (PDB: 6w4h, 6w61, 6w75, 6wjt, 6wkq, 6wks, 6wq3, 6wrz, 6wvn, 6xkm, 6yz1, 7bq7, 7c2i, 7c2j, 7diy, 7egq, 7eiz, 7jhe, 7jib, 7jpe, 7jyy, 7jz0, 7koa, 7l6r, 7l6t, 7lw3, 7lw4, 7mb9, 7mc5, 7mc6, 7n0b, 7n0c, 7n0d, 7r1t, 7r1u, 7ult, 8a23, 8bsd, 8bzv, 8c5m, 8f4s, 8f4y, 8osx, 8ot0, 8oto, 8otr, 8ov1, 8ov2, 8ov3, 8ov4, 8rv4, 8rv5, 8rv6, 8rv7, 8rv8, 8rv9, 8rva, 8rvb, 8rzc, 8rzd, 8rze, 8s8w, 8s8x, 8tyj, 8vuo, 9emj, 9eml, 9emv, 9eun, 9fw2, 9fwh, 9fwi, 9fwj, 9fwk, 9fwl, 9fwm, 9fwn, 9fwo, 9fwp, 9fwq, 9fwr, 9fws, 9fwt, 9fwu, 9fz4, 9fzk, 9gny, 9grp, 9grq, 9gs4, 9gtf, 9gud, 9gue, 9guf, 9guy, 9gwo, 9p6p, 9q1j, 9q8h, 9vck, 9vcl, 9yrk, 9yrl, 9yrn, 9yro) exist and high quality models can be extracted from them but should be used with care.
High quality models are available which are predicted to form homo-dodecamers.
>sp|P0DTD1|R1AB_SARS2|4393-5324 SADAQSFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNDKVAGFAKFLKTNCCRFQEKD EDDNLIDSYFVVKRHTFSNYQHEETIYNLLKDCPAVAKHDFFKFRIDGDMVPHISRQRLT KYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKKDWYDFVENPDILRVYANLGE RVRQALLKTVQFCDAMRNAGIVGVLTLDNQDLNGNWYDFGDFIQTTPGSGVPVVDSYYSL LMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERLKLFDRYFKYWDQTYHPNCVN CLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVN LHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAALTNNVAFQTVKPGNFNKDFY DFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYF DCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTIT QMNLKYAISAKNRARTVAGVSICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHN MLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLARKHTTCCSLSHRFYRLANECA QVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYV RNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSDDAVVCFNSTYASQGLVASIK NFKSVLYYQNNVFMSEAKCWTETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGA GCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADVFHLYLQYIRKLHDELTGHML DMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQ
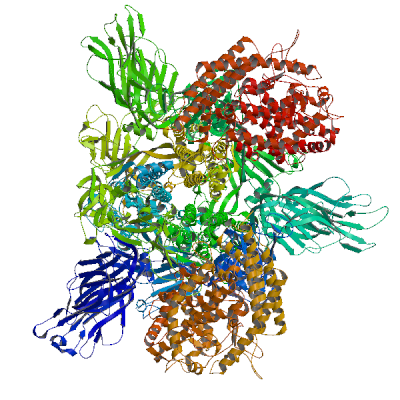
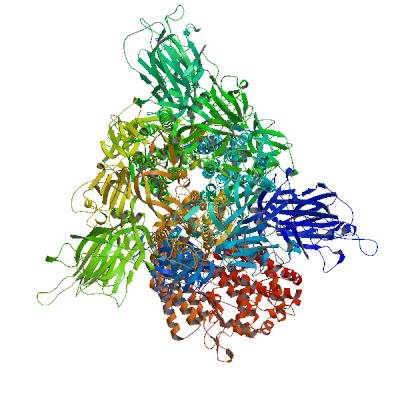
RNA-directed RNA polymerase nsp12 (Pol)
RNA-directed RNA polymerase that catalyzes the transcription of viral genomic and subgenomic RNAs. Acts in complex with nsp7 and nsp8 to transcribe both the minus and positive strands of genomic RNA (PubMed:32277040, PubMed:32358203, PubMed:32438371, PubMed:32526208). The kinase-like NiRAN domain of NSP12 attaches one or more nucleotides to the amino terminus of NSP9, forming a covalent RNA-protein intermediate that serves as transcription/replication primer (PubMed:37794589). Subgenomic RNAs (sgRNAs) are formed by discontinuous transcription: The polymerase has the ability to pause at transcription-regulating sequences (TRS) and jump to the leader TRS, resulting in a major deletion (PubMed:35706445). This creates a series of subgenomic RNAs that are replicated, transcribed and translated (PubMed:35706445). In addition, Nsp12 is a subunit of the viral RNA capping enzyme that catalyzes the RNA guanylyltransferase reaction for genomic and sub-genomic RNAs (PubMed:35944563). Subsequently, the NiRAN domain transfers RNA to GDP, and forms the core cap structure GpppA-RNA (PubMed:35944563).
Experimental structures of hetero-oligomeric complexes (PDB: 6m71, 6xez, 6xqb, 6yyt, 7aap, 7b3b, 7b3c, 7b3d, 7btf, 7bv1, 7bv2, 7bw4, 7bzf, 7c2k, 7ctt, 7cxm, 7cxn, 7cyq, 7d4f, 7dfg, 7dfh, 7doi, 7dok, 7dte, 7ed5, 7egq, 7eiz, 7krn, 7kro, 7krp, 7l1f, 7o7y, 7o7z, 7o80, 7o81, 7oyg, 7ozu, 7ozv, 7rdx, 7rdy, 7rdz, 7re0, 7re1, 7re2, 7re3, 7thm, 7uo4, 7uo7, 7uo9, 7uob, 7uoe, 8gw1, 8gwb, 8gwe, 8gwf, 8gwg, 8gwi, 8gwk, 8gwm, 8gwn, 8gwo, 8gy6, 8sq9, 8sqj, 8sqk, 8xch, 8xko, 9blf, 9cgv, 9dj8, 9i81, 9ikz, 9imk, 9imm, 9l09, 9pyw, 9pyz, 9pz0, 9sao, 9sap, 9saq, 9sar, 9uht) exist and high quality models can be extracted from them but should be used with care.
>sp|P0DTD1|R1AB_SARS2|5325-5925 AVGACVLCNSQTSLRCGACIRRPFLCCKCCYDHVISTSHKLVLSVNPYVCNAPGCDVTDV TQLYLGGMSYYCKSHKPPISFPLCANGQVFGLYKNTCVGSDNVTDFNAIATCDWTNAGDY ILANTCTERLKLFAAETLKATEETFKLSYGIATVREVLSDRELHLSWEVGKPRPPLNRNY VFTGYRVTKNSKVQIGEYTFEKGDYGDAVVYRGTTTYKLNVGDYFVLTSHTVMPLSAPTL VPQEHYVRITGLYPTLNISDEFSSNVANYQKVGMQKYSTLQGPPGTGKSHFAIGLALYYP SARIVYTACSHAAVDALCEKALKYLPIDKCSRIIPARARVECFDKFKVNSTLEQYVFCTV NALPETTADIVVFDEISMATNYDLSVVNARLRAKHYVYIGDPAQLPAPRTLLTKGTLEPE YFNSVCRLMKTIGPDMFLGTCRRCPAEIVDTVSALVYDNKLKAHKDKSAQCFKMFYKGVI THDVSSAINRPQIGVVREFLTRNPAWRKAVFISPYNSQNAVASKILGLPTQTVDSSQGSE YDYVIFTQTTETAHSCNVNRFNVAITRAKVGILCIMSDRDLYDKLQFTSLEIPRRNVATL Q
Helicase nsp13 (Hel)
Plays a role in viral RNA synthesis (PubMed:33232691). Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5' to 3' polarity. Activity of helicase is dependent on magnesium (By similarity). Binds to host TBK1 and inhibits TBK1 phosphorylation; the interaction with TBK1 decreases IRF3 phosphorylation, which leads to reduced IFN-beta production (PubMed:32979938).
Experimental structures (PDB: 5rl6, 5rl7, 5rl8, 5rl9, 5rlb, 5rlc, 5rld, 5rle, 5rlf, 5rlg, 5rlh, 5rli, 5rlj, 5rlk, 5rll, 5rlm, 5rln, 5rlo, 5rlp, 5rlq, 5rlr, 5rls, 5rlt, 5rlu, 5rlv, 5rlw, 5rly, 5rlz, 5rm0, 5rm1, 5rm2, 5rm3, 5rm4, 5rm5, 5rm6, 5rm7, 5rm8, 5rm9, 5rma, 5rmb, 5rmc, 5rmd, 5rme, 5rmf, 5rmg, 5rmh, 5rmi, 5rmj, 5rmk, 5rml, 5rmm, 5rob, 6zsl, 7nio, 7nn0, 7nng, 9i1s, 9i4v, 9i51, 9i53) are available. Experimental structures of hetero-oligomeric complexes (PDB: 6xez, 7cxm, 7cxn, 7cyq, 7egq, 7eiz, 7krn, 7kro, 7lfz, 7rdx, 7rdy, 7rdz, 7re0, 7re1, 7re2, 7re3, 8gw1, 8gwb, 8gwe, 8gwf, 8gwg, 8gwi, 8gwk, 8gwm, 8gwn, 8gwo, 8rne, 8rnf, 8xch, 9ikz, 9imk, 9imm, 9uht) exist and high quality models can be extracted from them but should be used with care.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|5926-6452 AENVTGLFKDCSKVITGLHPTQAPTHLSVDTKFKTEGLCVDIPGIPKDMTYRRLISMMGF KMNYQVNGYPNMFITREEAIRHVRAWIGFDVEGCHATREAVGTNLPLQLGFSTGVNLVAV PTGYVDTPNNTDFSRVSAKPPPGDQFKHLIPLMYKGLPWNVVRIKIVQMLSDTLKNLSDR VVFVLWAHGFELTSMKYFVKIGPERTCCLCDRRATCFSTASDTYACWHHSIGFDYVYNPF MIDVQQWGFTGNLQSNHDLYCQVHGNAHVASCDAIMTRCLAVHECFVKRVDWTIEYPIIG DELKINAACRKVQHMVVKAALLADKFPVLHDIGNPKAIKCVPQADVEWKFYDAQPCSDKA YKIEELFYSYATHSDKFTDGVCLFWNCNVDRYPANSIVCRFDTRVLSNLNLPGCDGGSLY VNKHAFHTPAFDKSAFVNLKQLPFFYYSDSPCESHGKQVVSDIDYVPLKSATCITRCNLG GAVCRHHANEYRLYLDAYNMMISAGFSLWVYKQFDTYNLWNTFTRLQ
Guanine-N7 methyltransferase nsp14
Plays a role in viral RNA synthesis through two distinct activities. The N7-guanine methyltransferase activity plays a role in the formation of the cap structure GpppA-RNA (PubMed:35944563). The proofreading exoribonuclease reduces the sensitivity of the virus to RNA mutagens during replication (By similarity). This activity acts on both ssRNA and dsRNA in a 3'-5' direction (By similarity).
Experimental structures (PDB: 5skw, 5skx, 5sky, 5skz, 5sl0, 5sl1, 5sl2, 5sl3, 5sl4, 5sl5, 5sl6, 5sl7, 5sl8, 5sl9, 5sla, 5slb, 5slc, 5sld, 5sle, 5slf, 5slg, 5slh, 5sli, 5slj, 5slk, 5sll, 5slm, 5sln, 5slo, 5slp, 5slq, 5slr, 5sls, 5slt, 5slu, 5slv, 5slw, 5slx, 5sly, 5slz, 5sm0, 5sm1, 5sm2, 5sm3, 5sm4, 5sm5, 5sm6, 5sm7, 5sm8, 5sm9, 5sma, 5smb, 5smc, 5smd, 5sme, 5smf, 5smg, 5smh, 5smi, 5smk, 7qgi, 7qif, 7r2v, 7tw7, 7tw8, 7tw9, 8bwu, 8frj, 8frk, 9feh, 9naz, 9nfp, 9nha, 9nhu, 9nio, 9njg, 9s0m, 9saj, 9sak, 9sal, 9sam, 9san, 9th6) are available. Experimental structures of hetero-oligomeric complexes (PDB: 7diy, 7egq, 7eiz, 7mc5, 7mc6, 7n0b, 7n0c, 7n0d, 8cmg, 9fw2, 9fwh, 9fwi, 9fwj, 9fwk, 9fwl, 9fwm, 9fwn, 9fwo, 9fwp, 9fwq, 9fwr, 9fws, 9fwt, 9fwu, 9fz4, 9fzk, 9q1j, 9vck, 9vcl, 9yrk, 9yrl, 9yrn, 9yro) exist and high quality models can be extracted from them but should be used with care.
High quality models can be extracted from hetero-oligomeric complexes but should be used with care.
>sp|P0DTD1|R1AB_SARS2|6453-6798 SLENVAFNVVNKGHFDGQQGEVPVSIINNTVYTKVDGVDVELFENKTTLPVNVAFELWAK RNIKPVPEVKILNNLGVDIAANTVIWDYKRDAPAHISTIGVCSMTDIAKKPTETICAPLT VFFDGRVDGQVDLFRNARNGVLITEGSVKGLQPSVGPKQASLNGVTLIGEAVKTQFNYYK KVDGVVQQLPETYFTQSRNLQEFKPRSQMEIDFLELAMDEFIERYKLEGYAFEHIVYGDF SHSQLGGLHLLIGLAKRFKESPFELEDFIPMDSTVKNYFITDAQTGSSKCVCSVIDLLLD DFVEIIKSQDLSVVSKVVKVTIDYTEISFMLWCKDGHVETFYPKLQ
Uridylate-specific endoribonuclease nsp15
Plays a role in viral transcription/replication and prevents the simultaneous activation of host cell dsRNA sensors, such as MDA5/IFIH1, OAS, and PKR (By similarity). Acts by degrading the 5'-polyuridines generated during replication of the poly(A) region of viral genomic and subgenomic RNAs (PubMed:33504779, PubMed:33564093). Catalyzes a two-step reaction in which a 2'3'-cyclic phosphate (2'3'-cP) is first generated by 2'-O transesterification, which is then hydrolyzed to a 3'-phosphate (3'-P) (PubMed:33504779, PubMed:33564093). If not degraded, poly(U) RNA would hybridize with poly(A) RNA tails and activate host dsRNA sensors (By similarity). May bind genomic dsRNA in association with the replication-transcription complex (RTC), and play a role in nsp12 discontinous transcription (PubMed:34562452, PubMed:35706445).
Experimental structures (PDB: 5s6x, 5s6y, 5s6z, 5s70, 5s71, 5s72, 5sa4, 5sa5, 5sa6, 5sa7, 5sa8, 5sa9, 5saa, 5sab, 5sac, 5sad, 5sae, 5saf, 5sag, 5sah, 5sai, 5sbf, 6vww, 6w01, 6wlc, 6wxc, 6x1b, 6x4i, 6xdh, 7k0r, 7k1l, 7k1o, 7k9p, 7keg, 7keh, 7kf4, 7me0, 7n06, 7n33, 7n7r, 7n7u, 7n7w, 7n7y, 7n83, 7rb0, 7rb2, 7tj2, 7tqv, 8d34, 8u2x, 8ud2, 8ud3, 8ud4, 8ud5, 9bih, 9m48, 9m49, 9mru, 9mrw, 9mry) are available. Experimental structures of hetero-oligomeric complexes (PDB: 9hh5, 9hh6) exist and high quality models can be extracted from them but should be used with care.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|6799-7096 SSQAWQPGVAMPNLYKMQRMLLEKCDLQNYGDSATLPKGIMMNVAKYTQLCQYLNTLTLA VPYNMRVIHFGAGSDKGVAPGTAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVHT ANKWDLIISDMYDPKTKNVTKENDSKEGFFTYICGFIQQKLALGGSVAIKITEHSWNADL YKLMGHFAWWTAFVTNVNASSSEAFLIGCNYLGKPREQIDGYVMHANYIFWRNTNPIQLS SYSLFDMSKFPLKLRGTAVMSLKEGQINDMILSLLSKGRLIIRENNRVVISSDVLVNN
2'-O-methyltransferase nsp16
Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs (PubMed:35944563). N7-methyl guanosine cap is a prerequisite for binding of nsp16 (PubMed:35944563). Therefore, it plays an essential role in cap methylation of viral mRNAs, which is essential to evade the immune system, especially when restricted by human IFIT1 and IFIT3 (PubMed:35944563, PubMed:36722972, PubMed:36285486). May disrupt host mRNA splicing in nucleus by interacting with pre-mRNA Recognition Domains of the U1 and U2 snRNAs (PubMed:33080218).
Experimental structures of hetero-oligomeric complexes (PDB: 6w4h, 6w61, 6w75, 6wjt, 6wkq, 6wks, 6wq3, 6wrz, 6wvn, 6xkm, 6yz1, 7bq7, 7c2i, 7c2j, 7jhe, 7jib, 7jpe, 7jyy, 7jz0, 7koa, 7l6r, 7l6t, 7lw3, 7lw4, 7r1t, 7r1u, 7ult, 8a23, 8bsd, 8bzv, 8c5m, 8f4s, 8f4y, 8osx, 8ot0, 8oto, 8otr, 8ov1, 8ov2, 8ov3, 8ov4, 8rv4, 8rv5, 8rv6, 8rv7, 8rv8, 8rv9, 8rva, 8rvb, 8rzc, 8rzd, 8rze, 8s8w, 8s8x, 8tyj, 8vuo, 9emj, 9eml, 9emv, 9eun, 9gny, 9grp, 9grq, 9gs4, 9gtf, 9gud, 9gue, 9guf, 9guy, 9gwo, 9p6p, 9q8h) exist and high quality models can be extracted from them but should be used with care.
High quality models are available.
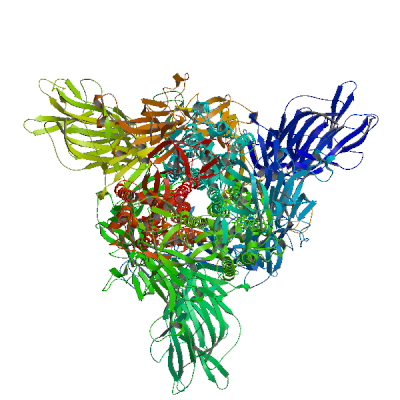
(1273 residues)
>sp|P0DTC2|SPIKE_SARS2 MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFS NVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIV NNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLE GKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQT LLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETK CTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISN CVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIAD YNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPC NGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVN FNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITP GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSY ECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTI SVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQE VFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDC LGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALN TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPA ICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDP LQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDL QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDD SEPVLKGVKLHYT
Spike glycoprotein (S glycoprotein)
Spike protein S1 (14-685): Attaches the virion to the cell membrane by interacting with host receptor, initiating the infection. The major receptor is host ACE2 (PubMed:32142651, PubMed:32155444, PubMed:33607086). When S2/S2' has been cleaved, binding to the receptor triggers direct fusion at the cell membrane (PubMed:34561887). When S2/S2' has not been cleaved, binding to the receptor results in internalization of the virus by endocytosis using host TFRC and GRM2 and leading to fusion of the virion membrane with the host endosomal membrane (PubMed:32075877, PubMed:32221306, PubMed:34903715, PubMed:36779763). Alternatively, may use NRP1/NRP2 (PubMed:33082294, PubMed:33082293) and integrin as entry receptors (PubMed:35150743). The use of NRP1/NRP2 receptors may explain the tropism of the virus in human olfactory epithelial cells, which express these molecules at high levels but ACE2 at low levels (PubMed:33082293). Uses also ASGR1 as an alternative receptor in an ACE2-independent manner (PubMed:34837059). The stalk domain of S contains three hinges, giving the head unexpected orientational freedom (PubMed:32817270). Spike protein S2 (686-1273): Precursor of the fusion protein processed in the biosynthesis of the S protein and the formation of virus particle. Mediates fusion of the virion and cellular membranes by functioning as a class I viral fusion protein. Contains two viral fusion peptides that are unmasked after cleavage. The S2/S2' cleavage occurs during virus entry at the cell membrane by host TMPRSS2 (PubMed:32142651) or during endocytosis by host CSTL (PubMed:32703818, PubMed:34159616). In either case, this triggers an extensive and irreversible conformational change leading to fusion of the viral envelope with the cellular cytoplasmic membrane, releasing viral genomic RNA into the host cell cytoplasm (PubMed:34561887). Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During fusion of the viral and target cell membranes, the coiled coil regions (heptad repeats) adopt a trimer-of-hairpins structure and position the fusion peptide in close proximity to the C-terminal region of the ectodomain. Formation of this structure appears to promote apposition and subsequent fusion of viral and target cell membranes. Spike protein S2' (816-1273): Subunit of the fusion protein that is processed upon entry into the host cell. Mediates fusion of the virion and cellular membranes by functioning as a class I viral fusion protein. Contains a viral fusion peptide that is unmasked after S2 cleavage. This cleavage can occur at the cell membrane by host TMPRSS2 or during endocytosis by host CSTL (PubMed:32703818, PubMed:34159616). In either case, this triggers an extensive and irreversible conformational change that leads to fusion of the viral envelope with the cellular cytoplasmic membrane, releasing viral genomic RNA into the host cell cytoplasm (PubMed:34561887). Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During fusion of the viral and target cell membranes, the coiled coil regions (heptad repeats) adopt a trimer-of-hairpins structure and position the fusion peptide in close proximity to the C-terminal region of the ectodomain. Formation of this structure appears to promote apposition and subsequent fusion of viral and target cell membranes.
Experimental structures (PDB: 6lvn, 6lxt, 6m1v, 6vsb, 6vxx, 6vyb, 6w41, 6wps, 6wpt, 6x29, 6x2a, 6x2b, 6x2c, 6x6p, 6x79, 6xc2, 6xc3, 6xc4, 6xc7, 6xcm, 6xcn, 6xdg, 6xe1, 6xf5, 6xf6, 6xkl, 6xkp, 6xkq, 6xlu, 6xm0, 6xm3, 6xm4, 6xm5, 6xr8, 6xra, 6xs6, 6yla, 6ym0, 6yor, 6yz5, 6yz7, 6z2m, 6z43, 6z97, 6zb4, 6zb5, 6zbp, 6zcz, 6zdg, 6zdh, 6zer, 6zfo, 6zge, 6zgg, 6zgh, 6zgi, 6zh9, 6zhd, 6zlr, 6zow, 6zox, 6zoy, 6zoz, 6zp0, 6zp1, 6zp2, 6zp5, 6zp7, 6zwv, 6zxn, 7a25, 7a29, 7a4n, 7a5r, 7a5s, 7a93, 7ad1, 7akd, 7b14, 7b17, 7b18, 7b27, 7b3o, 7b62, 7beh, 7bei, 7bej, 7bek, 7bel, 7bem, 7ben, 7beo, 7bep, 7bnm, 7bnn, 7bno, 7bnv, 7bwj, 7byr, 7bz5, 7c01, 7c2l, 7c8v, 7c8w, 7cab, 7cac, 7cah, 7cai, 7cak, 7can, 7cdi, 7cdj, 7ch4, 7ch5, 7chb, 7chc, 7che, 7chf, 7chh, 7cho, 7chp, 7chs, 7cjf, 7cm4, 7cn9, 7cot, 7cwl, 7cwm, 7cwn, 7cwo, 7cws, 7cwt, 7cwu, 7cyh, 7cyp, 7cyv, 7d0b, 7d0c, 7d0d, 7d2z, 7d30, 7d4g, 7d6i, 7dcc, 7dcx, 7dd2, 7dd8, 7ddd, 7ddn, 7deo, 7det, 7deu, 7df3, 7djz, 7dk0, 7dk2, 7dk3, 7dk4, 7dk5, 7dk6, 7dk7, 7dpm, 7dwy, 7dwz, 7dx0, 7dx1, 7dx2, 7dx3, 7dzw, 7dzx, 7dzy, 7e23, 7e39, 7e3b, 7e3c, 7e3j, 7e3k, 7e3l, 7e3o, 7e5o, 7e5r, 7e5s, 7e5y, 7e7b, 7e7d, 7e7x, 7e7y, 7e86, 7e88, 7e8c, 7e8m, 7e9n, 7e9o, 7e9p, 7e9q, 7e9t, 7eam, 7ean, 7eaz, 7eb0, 7eb3, 7eb4, 7eb5, 7edf, 7edg, 7edh, 7edi, 7eh5, 7ej4, 7ej5, 7ejy, 7ejz, 7ek0, 7enf, 7eng, 7epx, 7ey0, 7ey5, 7ezv, 7f0x, 7f12, 7f15, 7f3q, 7f46, 7f5g, 7f5h, 7f62, 7f63, 7f6y, 7f6z, 7f7e, 7f7h, 7fae, 7faf, 7fat, 7fau, 7fb0, 7fb1, 7fb3, 7fb4, 7fbj, 7fbk, 7fcd, 7fce, 7fcp, 7fcq, 7fet, 7fg2, 7fg3, 7fg7, 7fjc, 7fjn, 7fjo, 7fjs, 7jji, 7jjj, 7jmo, 7jmp, 7jmw, 7jv2, 7jv4, 7jv6, 7jva, 7jvb, 7jvc, 7jw0, 7jwb, 7jwy, 7jx3, 7jzl, 7jzm, 7jzn, 7jzu, 7k43, 7k45, 7k4n, 7k8m, 7k8s, 7k8t, 7k8u, 7k8v, 7k8w, 7k8x, 7k8y, 7k8z, 7k90, 7k9h, 7k9i, 7k9j, 7k9k, 7k9z, 7kdg, 7kdh, 7kdi, 7kdj, 7kdk, 7kdl, 7ke4, 7ke6, 7ke7, 7ke8, 7ke9, 7kea, 7keb, 7kec, 7kfv, 7kfw, 7kfx, 7kfy, 7kgj, 7kgk, 7kj5, 7kkk, 7kkl, 7kl9, 7klg, 7klh, 7klw, 7km5, 7kmg, 7kmh, 7kmi, 7kmk, 7kml, 7kn3, 7kn4, 7kn5, 7kn6, 7kn7, 7kqb, 7kqe, 7krq, 7krr, 7krs, 7ks9, 7ksg, 7kxj, 7kxk, 7kzb, 7l02, 7l06, 7l09, 7l2c, 7l2d, 7l2e, 7l2f, 7l3n, 7l4z, 7l56, 7l57, 7l58, 7l5b, 7l7d, 7l7e, 7l7k, 7laa, 7lab, 7lcn, 7ld1, 7ldj, 7ljr, 7lm8, 7lop, 7lq7, 7lqv, 7lqw, 7lrs, 7lrt, 7ls9, 7lss, 7lwi, 7lwj, 7lwk, 7lwl, 7lwm, 7lwn, 7lwo, 7lwp, 7lwq, 7lws, 7lwt, 7lwu, 7lwv, 7lww, 7lx5, 7lxw, 7lxx, 7lxy, 7lxz, 7ly0, 7ly2, 7ly3, 7lyk, 7lyl, 7lym, 7lyn, 7lyo, 7lyp, 7lyq, 7m0j, 7m3i, 7m42, 7m53, 7m6d, 7m6e, 7m6f, 7m6g, 7m6h, 7m6i, 7m71, 7m7b, 7m7w, 7m8j, 7m8k, 7mdw, 7me7, 7mej, 7mf1, 7mfu, 7mjg, 7mjh, 7mji, 7mjj, 7mjk, 7mjl, 7mkl, 7mkm, 7mlz, 7mm0, 7mmo, 7msq, 7mtc, 7mtd, 7mte, 7mw2, 7mw3, 7mw4, 7mw5, 7mw6, 7mxp, 7my2, 7my3, 7my8, 7mzf, 7mzg, 7mzh, 7mzi, 7mzj, 7mzk, 7mzl, 7mzm, 7mzn, 7n0g, 7n0h, 7n1q, 7n1t, 7n1u, 7n1v, 7n1w, 7n1x, 7n1y, 7n3i, 7n4i, 7n4j, 7n4l, 7n4m, 7n5h, 7n62, 7n64, 7n8h, 7n8i, 7n9a, 7n9b, 7n9c, 7n9e, 7n9t, 7nab, 7nd3, 7nd4, 7nd5, 7nd6, 7nd7, 7nd8, 7nd9, 7nda, 7ndb, 7ndc, 7ndd, 7neg, 7neh, 7nkt, 7nll, 7np1, 7ns6, 7nt9, 7nta, 7ntc, 7nx6, 7nx7, 7nx8, 7nx9, 7nxa, 7nxb, 7oan, 7oao, 7oap, 7oaq, 7oau, 7oay, 7od3, 7odl, 7olz, 7or9, 7ora, 7orb, 7owx, 7p40, 7p5g, 7p5q, 7p5s, 7p77, 7p78, 7p79, 7p7a, 7p7b, 7phg, 7pqy, 7pqz, 7pr0, 7pry, 7prz, 7ps0, 7ps1, 7ps2, 7ps4, 7ps5, 7ps6, 7ps7, 7q0a, 7q0g, 7q0h, 7q0i, 7q1z, 7q3q, 7q3r, 7q6e, 7q9f, 7q9g, 7q9i, 7q9j, 7q9k, 7q9m, 7q9p, 7qdg, 7qdh, 7qez, 7qf0, 7qf1, 7qnw, 7qnx, 7qny, 7qo7, 7qo9, 7qti, 7qtj, 7qtk, 7qur, 7qus, 7r13, 7r14, 7r15, 7r16, 7r17, 7r18, 7r19, 7r1b, 7r40, 7r4i, 7r4q, 7r4r, 7r6w, 7r6x, 7r7n, 7r8l, 7r8m, 7r8n, 7r8o, 7r95, 7ra8, 7ral, 7raq, 7rbv, 7rby, 7rku, 7rkv, 7rnj, 7rq6, 7rr0, 7ru1, 7ru2, 7ru3, 7ru4, 7ru5, 7ru8, 7rw2, 7s0b, 7s0c, 7s0d, 7s0e, 7s3n, 7s4s, 7s5p, 7s5q, 7s5r, 7s6i, 7s6j, 7s6k, 7s6l, 7s83, 7sbk, 7sbl, 7sbo, 7sbp, 7sbq, 7sbr, 7sbs, 7sbt, 7sbu, 7sc1, 7sd5, 7si2, 7sj0, 7sjs, 7skz, 7sl5, 7sn2, 7sn3, 7so9, 7soa, 7sob, 7soc, 7sod, 7soe, 7sof, 7spo, 7spp, 7swn, 7swo, 7swp, 7sww, 7swx, 7sxr, 7sxs, 7sxt, 7sxu, 7sxv, 7sxw, 7t01, 7t3m, 7t67, 7t72, 7t7b, 7t9j, 7tas, 7tat, 7tb4, 7tb8, 7tbf, 7tca, 7tcc, 7tei, 7tey, 7tf1, 7tf2, 7tf3, 7tf4, 7tf5, 7tf8, 7tge, 7tgw, 7tgx, 7tgy, 7the, 7thk, 7tht, 7tl1, 7tl9, 7tla, 7tlb, 7tlc, 7tld, 7tly, 7tm0, 7tnw, 7to4, 7tou, 7tov, 7tow, 7tox, 7toy, 7toz, 7tp0, 7tp1, 7tp2, 7tp3, 7tp4, 7tp7, 7tp8, 7tp9, 7tpa, 7tpc, 7tpe, 7tpf, 7tph, 7tpi, 7tpk, 7tpl, 7tpr, 7tyz, 7tz0, 7u09, 7u0a, 7u0d, 7u0e, 7u0p, 7u0q, 7u0x, 7u2d, 7u2e, 7u8e, 7u9o, 7u9p, 7uap, 7uaq, 7uar, 7ub0, 7ub5, 7ub6, 7uhc, 7ukl, 7ukm, 7ul0, 7upl, 7urq, 7urs, 7uz4, 7uz5, 7uz6, 7uz7, 7uz8, 7uz9, 7uza, 7uzb, 7uzc, 7uzd, 7v20, 7v22, 7v23, 7v24, 7v26, 7v27, 7v2a, 7v76, 7v77, 7v78, 7v79, 7v7a, 7v7d, 7v7e, 7v7f, 7v7g, 7v7h, 7v7i, 7v7j, 7v7n, 7v7o, 7v7p, 7v7q, 7v7r, 7v7s, 7v7t, 7v7u, 7v7v, 7v8c, 7vhh, 7vhj, 7vhk, 7vhl, 7vhm, 7vhn, 7vmu, 7vnb, 7vnc, 7vnd, 7vne, 7voa, 7vq0, 7vrv, 7vrw, 7vx1, 7vxe, 7vxi, 7vyr, 7vzt, 7w1s, 7w92, 7w94, 7w9e, 7w9f, 7wb5, 7wbz, 7wcd, 7wch, 7wck, 7wcp, 7wcr, 7wcu, 7wcz, 7wd0, 7wd1, 7wd2, 7wd7, 7wd8, 7wd9, 7wdf, 7we7, 7we8, 7we9, 7wea, 7web, 7wec, 7wed, 7wee, 7wef, 7wev, 7wg6, 7wg7, 7wg8, 7wg9, 7wgv, 7wgx, 7wgy, 7wgz, 7wh8, 7whb, 7whd, 7whi, 7whj, 7whk, 7wjy, 7wjz, 7wk0, 7wk2, 7wk3, 7wk8, 7wk9, 7wka, 7wlc, 7wly, 7wlz, 7wm0, 7wn2, 7wnb, 7wo4, 7wo5, 7wo7, 7woa, 7wob, 7woc, 7wog, 7wop, 7woq, 7wor, 7wos, 7wou, 7wov, 7wow, 7wp6, 7wp8, 7wp9, 7wpd, 7wpe, 7wpf, 7wph, 7wqv, 7wr8, 7wrj, 7wrv, 7ws0, 7ws1, 7ws2, 7ws3, 7ws4, 7ws5, 7ws6, 7ws7, 7wse, 7wt7, 7wt8, 7wt9, 7wtf, 7wtg, 7wth, 7wti, 7wtj, 7wtk, 7wue, 7wuh, 7wvl, 7wwi, 7wwj, 7wwk, 7wwl, 7wwm, 7wz1, 7wz2, 7x1m, 7x2h, 7x2k, 7x2l, 7x2m, 7x63, 7x66, 7x6a, 7x7d, 7x7e, 7x7n, 7x7o, 7x7t, 7x7u, 7x8w, 7x8y, 7x8z, 7x90, 7x91, 7x92, 7x93, 7x94, 7x95, 7x96, 7x9e, 7xck, 7xco, 7xcz, 7xd2, 7xda, 7xdb, 7xdk, 7xdl, 7xeg, 7xei, 7xh8, 7xic, 7xik, 7xil, 7xiw, 7xix, 7xiy, 7xiz, 7xj6, 7xj8, 7xj9, 7xmx, 7xmz, 7xnq, 7xnr, 7xns, 7xod, 7xs8, 7xsa, 7xsb, 7xsc, 7xst, 7xtz, 7xu0, 7xu1, 7xu2, 7xu3, 7xu4, 7xu5, 7xu6, 7xy3, 7xy4, 7y0c, 7y0n, 7y0o, 7y0v, 7y3o, 7y42, 7y6d, 7y6k, 7y6l, 7y6n, 7y7j, 7y7k, 7y9s, 7yad, 7ybh, 7ybi, 7ybj, 7ybk, 7ybl, 7ybm, 7ybn, 7yc5, 7yck, 7ycl, 7ycn, 7yco, 7yd1, 7ydy, 7ye5, 7ye9, 7yh6, 7yh7, 7ykj, 7yow, 7yqt, 7yqu, 7yqv, 7yqw, 7yqx, 7yqy, 7yqz, 7yr0, 7yr1, 7ytn, 7yue, 7yve, 7yvf, 7yvg, 7yvh, 7yvi, 7yvj, 7yvk, 7yvl, 7yvm, 7yvn, 7yvo, 7yvp, 7z0x, 7z0y, 7z1a, 7z1b, 7z1c, 7z1d, 7z1e, 7z3z, 7z6v, 7z7x, 7z85, 7z86, 7z8o, 7z9q, 7z9r, 7zbu, 7zce, 7zcf, 7zf3, 7zf4, 7zf5, 7zf8, 7zf9, 7zfa, 7zfb, 7zfc, 7zfd, 7zfe, 7zjl, 7zr7, 7zr8, 7zr9, 7zrc, 7zxu, 8a99, 8aaa, 8bbn, 8bcz, 8be1, 8bec, 8bev, 8bg2, 8bg3, 8bg4, 8bg5, 8bg6, 8bg8, 8bgg, 8bh5, 8bon, 8bse, 8bsf, 8c0y, 8c1v, 8c2r, 8c3v, 8c8p, 8cbd, 8cbe, 8cbf, 8cim, 8cin, 8cma, 8csa, 8csj, 8cwi, 8cwu, 8cwv, 8cxn, 8cxq, 8cy6, 8cy7, 8cy9, 8cya, 8cyb, 8cyc, 8cyd, 8cyj, 8d0z, 8d36, 8d47, 8d48, 8d55, 8d56, 8d5a, 8d6z, 8d8q, 8d8r, 8dad, 8dcc, 8dce, 8dgu, 8di5, 8dli, 8dll, 8dlo, 8dlr, 8dls, 8dlt, 8dlw, 8dlx, 8dly, 8dlz, 8dm0, 8dm1, 8dm2, 8dm3, 8dm4, 8dnn, 8dpz, 8dt3, 8dt8, 8dtk, 8dtr, 8dtt, 8dtx, 8dw2, 8dw3, 8dw9, 8dwa, 8dxs, 8dxt, 8dxu, 8dya, 8dzh, 8dzi, 8e1g, 8el2, 8elj, 8elo, 8elp, 8elq, 8eoo, 8epn, 8epp, 8epq, 8eqf, 8erq, 8err, 8eyg, 8eyh, 8f0g, 8f0h, 8f0i, 8f2j, 8f2x, 8f4p, 8fah, 8fez, 8fhy, 8fi9, 8fu7, 8fu8, 8fu9, 8g70, 8g71, 8g72, 8g73, 8g74, 8g75, 8g76, 8g77, 8g78, 8g79, 8g7a, 8g7b, 8g7c, 8gb0, 8gb5, 8gb6, 8gb7, 8gb8, 8gdr, 8gf2, 8gjm, 8gjn, 8gnh, 8gou, 8gpy, 8gs6, 8gs9, 8gsb, 8gto, 8gtp, 8gtq, 8gx9, 8gz5, 8gzz, 8h00, 8h01, 8h07, 8h08, 8h3d, 8h3e, 8h3m, 8h3n, 8h5t, 8h5u, 8h6f, 8h7l, 8h7z, 8h91, 8hc2, 8hc3, 8hc4, 8hc5, 8hc8, 8hc9, 8hca, 8hcb, 8heb, 8hec, 8hed, 8hes, 8hfx, 8hfy, 8hfz, 8hg0, 8hgl, 8hgm, 8hhx, 8hhy, 8hhz, 8hlc, 8hld, 8hn6, 8hn7, 8hp9, 8hpf, 8hpq, 8hpu, 8hpv, 8hq7, 8hr2, 8hrd, 8hri, 8hrj, 8hws, 8hwt, 8i3s, 8i3u, 8i4e, 8i4f, 8i4g, 8i4h, 8i5h, 8i5i, 8idn, 8ify, 8ifz, 8ios, 8iot, 8itu, 8iv4, 8iv5, 8iv8, 8iva, 8ix3, 8j1q, 8j1t, 8j1v, 8j26, 8jap, 8jin, 8jio, 8jmm, 8jva, 8jyk, 8jym, 8jys, 8k18, 8k19, 8k3k, 8k45, 8k46, 8k47, 8k5g, 8k5h, 8k9b, 8k9j, 8k9m, 8kdr, 8keh, 8kej, 8keo, 8kep, 8keq, 8ker, 8khc, 8khd, 8owt, 8owv, 8oww, 8oyt, 8oyu, 8p5m, 8p99, 8p9y, 8pq2, 8psd, 8q5y, 8q7s, 8q93, 8q94, 8q95, 8qpr, 8qq0, 8qrf, 8qrg, 8qtd, 8qzr, 8r1c, 8r80, 8r8k, 8rby, 8rj7, 8rrn, 8sdf, 8sdg, 8sdh, 8sgu, 8siq, 8sir, 8sis, 8sit, 8sk5, 8smi, 8smt, 8suo, 8swh, 8t21, 8thf, 8tm1, 8tma, 8tmy, 8tyl, 8tyo, 8u1g, 8u28, 8ug9, 8uir, 8uk1, 8ukd, 8ukf, 8upx, 8usz, 8uul, 8uum, 8uun, 8uuo, 8v0l, 8v0m, 8v0n, 8v0o, 8v0p, 8v0q, 8v0r, 8v0s, 8v0t, 8v0u, 8v0v, 8v0w, 8v0x, 8v4f, 8v5v, 8vao, 8vcr, 8vif, 8vke, 8vkk, 8vye, 8vyf, 8vyg, 8w4f, 8wfh, 8wfm, 8whv, 8whw, 8wmd, 8wmf, 8wpw, 8wpy, 8wxl, 8wyj, 8wzi, 8x0x, 8x0y, 8x4h, 8x4z, 8x50, 8x55, 8x56, 8x5q, 8x5r, 8xbf, 8xe9, 8xea, 8xef, 8xi6, 8xk2, 8xki, 8xlv, 8xm5, 8xmg, 8xmt, 8xrq, 8xsd, 8xse, 8xsi, 8xsl, 8xur, 8xus, 8xut, 8xuu, 8xux, 8y0y, 8y4a, 8y4c, 8y5j, 8ydp, 8ydr, 8yds, 8ydt, 8ydx, 8ydy, 8ydz, 8yk4, 8ykg, 8ykh, 8yro, 8yrp, 8yub, 8yuc, 8ywe, 8yww, 8ywx, 8yz5, 8yz6, 8z2e, 8z6q, 8z6r, 8z6s, 8z6t, 8z6u, 8z6w, 8z6x, 8z7g, 8z7l, 8z86, 8zby, 8zbz, 8zc0, 8zc1, 8zc2, 8zc3, 8zc4, 8zc5, 8zc6, 8zer, 8zes, 8zhd, 8zhe, 8zhf, 8zhg, 8zhh, 8zhi, 8zhj, 8zhk, 8zhl, 8zhm, 8zhn, 8zho, 8zhp, 8zpp, 8zpq, 8zrd, 8zyf, 9aru, 9ato, 9atp, 9atq, 9atr, 9au2, 9ayw, 9ayx, 9ayy, 9b0y, 9b2v, 9b4x, 9b82, 9b8f, 9b9u, 9bbk, 9bd9, 9bea, 9bj2, 9bj3, 9bj4, 9bll, 9c44, 9c45, 9c7s, 9c7x, 9cb0, 9cci, 9ccj, 9cfe, 9cff, 9cfg, 9cfh, 9co6, 9co7, 9co8, 9co9, 9cpp, 9cpq, 9cpr, 9cps, 9cpt, 9cpu, 9cpv, 9cpw, 9cpx, 9cpy, 9crc, 9crd, 9cre, 9crf, 9crg, 9crh, 9cri, 9css, 9ct2, 9cvh, 9cwp, 9cwq, 9cwr, 9cxe, 9d8h, 9d8i, 9d8j, 9d8k, 9d8l, 9dhy, 9e21, 9ecz, 9elh, 9eli, 9elj, 9elk, 9ell, 9elm, 9eln, 9elo, 9elp, 9elq, 9f9y, 9fc2, 9fcm, 9fgr, 9fgs, 9fgt, 9fgu, 9fjk, 9fmw, 9gdx, 9gdy, 9gxe, 9gxg, 9h6u, 9iqp, 9j66, 9jeb, 9js4, 9k6j, 9k6y, 9kt3, 9kvd, 9kve, 9kvf, 9kvj, 9kvk, 9kvq, 9kvt, 9kzd, 9kze, 9kzz, 9l05, 9l07, 9l15, 9l2l, 9l6c, 9lae, 9lbs, 9lds, 9lh2, 9loy, 9loz, 9lp0, 9lp1, 9ls3, 9lyo, 9lyp, 9mi3, 9ml4, 9ml5, 9ml6, 9ml7, 9ml8, 9ml9, 9mpw, 9mr1, 9mr2, 9nvg, 9nxy, 9o5t, 9o5v, 9og4, 9og5, 9og6, 9og7, 9psn, 9pso, 9psp, 9pw4, 9q3u, 9q3v, 9ssm, 9uel, 9uxd, 9uxe, 9y5y) are available. Experimental structures of hetero-oligomeric complexes (PDB: 6lzg, 6m0j, 6m17, 6vw1, 6x45, 6xey, 7a91, 7a92, 7a94, 7a95, 7a96, 7a97, 7a98, 7b0b, 7bh9, 7c8d, 7c8j, 7ct5, 7czp, 7czq, 7czr, 7czs, 7czt, 7czu, 7czv, 7czw, 7czx, 7czy, 7czz, 7d00, 7d03, 7df4, 7dhx, 7dmu, 7dqa, 7dwx, 7dx4, 7dx5, 7dx6, 7dx7, 7dx8, 7dx9, 7e8f, 7edj, 7efp, 7efr, 7ek6, 7ekc, 7eke, 7ekf, 7ekg, 7ekh, 7ey4, 7eya, 7f5r, 7fc5, 7fdg, 7fdh, 7fdi, 7fdk, 7fem, 7kj2, 7kj3, 7kj4, 7kmb, 7kms, 7kmz, 7knb, 7kne, 7knh, 7kni, 7l0n, 7l7f, 7lo4, 7mjm, 7mjn, 7nxc, 7p19, 7r0z, 7r10, 7r11, 7r12, 7r1a, 7rbu, 7rpv, 7rxd, 7rzq, 7rzr, 7rzs, 7rzt, 7rzu, 7rzv, 7sn0, 7sxx, 7sxy, 7sxz, 7sy0, 7sy1, 7sy2, 7sy3, 7sy4, 7sy5, 7sy6, 7sy7, 7sy8, 7t9k, 7t9l, 7tew, 7tex, 7tez, 7tf0, 7tik, 7tn0, 7u0n, 7ufk, 7ufl, 7v7z, 7v80, 7v81, 7v82, 7v83, 7v84, 7v85, 7v86, 7v87, 7v88, 7v89, 7v8a, 7v8b, 7vx4, 7vx5, 7vx9, 7vxa, 7vxb, 7vxc, 7vxd, 7vxf, 7vxk, 7vxm, 7w6u, 7w8s, 7w99, 7w9b, 7w9c, 7wa1, 7wbl, 7wbp, 7wbq, 7wgb, 7wgc, 7whh, 7whz, 7wk4, 7wk5, 7wk6, 7wnm, 7won, 7wp0, 7wp1, 7wp2, 7wp5, 7wpa, 7wpb, 7wpc, 7wrh, 7wri, 7ws8, 7ws9, 7wsa, 7wsh, 7wsk, 7wvp, 7wvq, 7xa7, 7xaz, 7xb0, 7xb1, 7xby, 7xch, 7xci, 7xcp, 7xid, 7xo4, 7xo5, 7xo6, 7xo7, 7xo8, 7xo9, 7xoa, 7xob, 7xoc, 7xrp, 7xwa, 7xxl, 7y1y, 7y1z, 7y20, 7y21, 7y75, 7y76, 7y9n, 7y9z, 7ya0, 7ya1, 7ydi, 7yeg, 7yhw, 7yj3, 7yr2, 7yr3, 7yr4, 7yv8, 7yvu, 7zdq, 7zf7, 7zr2, 7zrv, 7zsd, 7zss, 8aqs, 8aqt, 8aqu, 8aqv, 8aqw, 8bbo, 8cmb, 8cmc, 8cmd, 8cmi, 8cwk, 8czi, 8dao, 8df5, 8dlj, 8dlk, 8dlm, 8dln, 8dlp, 8dlq, 8dlu, 8dlv, 8dm5, 8dm6, 8dm7, 8dm8, 8dm9, 8dma, 8dv1, 8dv2, 8edf, 8fa1, 8fa2, 8gry, 8h06, 8h5c, 8hc6, 8hc7, 8hrk, 8hrl, 8i9b, 8i9c, 8i9d, 8i9e, 8i9f, 8i9g, 8i9h, 8if2, 8iou, 8iov, 8jje, 8jyn, 8jyo, 8jyp, 8ka8, 8kc2, 8qh0, 8qh1, 8qsq, 8sph, 8spi, 8t20, 8t22, 8t25, 8taz, 8uze, 8via, 8vkl, 8vkm, 8vkn, 8vko, 8vkp, 8vqr, 8vya, 8wdr, 8wds, 8wdy, 8wdz, 8we0, 8we1, 8we4, 8wgv, 8wgw, 8whs, 8whu, 8whz, 8wlo, 8wlr, 8wox, 8woy, 8wp8, 8wrh, 8wrl, 8wrm, 8wro, 8wtd, 8wtj, 8wyh, 8xal, 8xlm, 8xln, 8xn2, 8xn3, 8xn5, 8xnf, 8xnk, 8xsf, 8xsj, 8xuy, 8xuz, 8xv0, 8xv1, 8xvm, 8xxw, 8xy9, 8xye, 8xyg, 8xyo, 8xyz, 8xz0, 8xz4, 8xz5, 8xz8, 8xz9, 8xza, 8xzd, 8y16, 8y18, 8y32, 8y6a, 8yf2, 8yft, 8yzb, 8yzc, 8yzd, 8yze, 8z3w, 8z4x, 8z64, 8z6a, 8z7b, 8z7p, 8z9l, 8zbq, 9bnd, 9bne, 9bnf, 9bng, 9ele, 9elf, 9elg, 9iu1, 9iup, 9iuq, 9iuu, 9jrc, 9jte, 9kud, 9ue6, 9ue7, 9ug3, 9xgo) exist and high quality models can be extracted from them but should be used with care.
High quality models are available which are predicted to form homo-trimers.
(275 residues)
>sp|P0DTC3|AP3A_SARS2 MDLFMRIFTIGTVTLKQGEIKDATPSDFVRATATIPIQASLPFGWLIVGVALLAVFQSAS KIITLKKRWQLALSKGVHFVCNLLLLFVTVYSHLLLVAAGLEAPFLYLYALVYFLQSINF VRIIMRLWLCWKCRSKNPLLYDANYFLCWHTNCYDYCIPYNSVTSSIVITSGDGTTSPIS EHDYQIGGYTEKWESGVKDCVVLHSYFTSDYYQLYSTQLSTDTGVEHVTFFIYNKIVDEP EEHVQIHTIDGSSGVVNPVMEPIYDEPTTTTSVPL
ORF3a protein (ORF3a)
Plays a role in viral egress via lysosomal trafficking (PubMed:33157038, PubMed:33422265). Forms homotetrameric ion channels (viroporins) localized at endosomes and lysosomes, that may induce deacidification of lysosomes, allowing safe egress of virions via lysosomal trafficking (PubMed:33157038, PubMed:33422265, PubMed:34158638). Also blocks autolysosome formation by binding and sequestering the host component VPS39 for homotypic fusion and protein sorting (HOPS) on late endosomes (PubMed:33422265). This prevents fusion of autophagosomes with lysosomes, disrupting autophagy and facilitating virus egress (PubMed:33422265). Induces host RETREG1/FAM134B-dependent reticulophagy by interacting with host HMGB1 and enhancing the association between HMGB1 and host BECN1 (PubMed:35239449). This induces endoplasmic reticulum stress and inflammatory responses and facilitates viral infection (PubMed:35239449).
Experimental structures (PDB: 6xdc, 8eqj, 8eqt) are available. Experimental structures of hetero-oligomeric complexes (PDB: 7kjr, 8equ, 8t5p, 8t5q, 9ru5) exist and high quality models can be extracted from them but should be used with care.
High quality models are available which are predicted to form homo-dimers.
(22 residues)
>sp|P0DTF1|ORF3B_SARS2 MMPTIFFAGILIVTTIVYLTIV
Putative ORF3b protein (ORF3b)
Acts as an interferon antagonist when expressed ex vivo.
Sequence is too short for modelling.
(41 residues)
>sp|P0DTG1|ORF3C_SARS2 MLLLQILFALLQRYRYKPHSLSDGLLLALHFLLFFRALPKS
(57 residues)
>sp|P0DTG0|ORF3D_SARS2 MAYCWRCTSCCFSERFQNHNPQKEMATSTLQGCSLCLQLAVVVCNSLLTPFARCCWP
Putative ORF3d protein
Only remote homologues were identified as potential template structures, no models have been built.
See here for template search results.
(75 residues)
>sp|P0DTC4|VEMP_SARS2 MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTALRLCAYCCNIVNVSLVKPSFYVYS RVKNLNSSRVPDLLV
Envelope small membrane protein (E)
Plays a central role in virus morphogenesis and assembly. Acts as a viroporin and self-assembles in host membranes forming pentameric protein-lipid pores that allow ion transport. Also plays a role in the induction of apoptosis (By similarity). Regulates the localization of S protein at cis-Golgi, the place of virus budding (PubMed:33229438). May act by slowing down the cell secretory pathway (PubMed:33229438). May interfere with tight-junction stability by interacting with host MPP5. This would result in disruption of epithelial barriers, thereby amplifying inflammatory processes (PubMed:32891874).
Partial coverage by experimental structures (PDB: 7k3g, 8suz, 8u1t) is available. Partial coverage by experimental structures of hetero-oligomeric complexes exist (PDB: 7m4r, 7ntk, 7qcr, 7qcs, 7qct, 7tuq, 7tv0, 9hm2) and high quality models can be extracted from them but should be used with care.
Model quality estimates are low - use with care.
(222 residues)
>sp|P0DTC5|VME1_SARS2 MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYANRNRFLYIIKLIFLWLLWPV TLACFVLAAVYRINWITGGIAIAMACLVGLMWLSYFIASFRLFARTRSMWSFNPETNILL NVPLHGTILTRPLLESELVIGAVILRGHLRIAGHHLGRCDIKDLPKEITVATSRTLSYYK LGASQRVAGDSGFAAYSRYRIGNYKLNTDHSSSSDNIALLVQ
Membrane protein (M)
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins (By similarity). Regulates the localization of S protein at cis-Golgi, the place of virus budding (PubMed:33229438). May act by binding cytoplasmic c-terminus of S (PubMed:33229438).
Experimental structures (PDB: 7vgr, 7vgs, 8ctk, 9ctu, 9ctw, 9exa) are available. Partial coverage by experimental structures of hetero-oligomeric complexes exist (PDB: 8cme, 9bf9) and high quality models can be extracted from them but should be used with care.
Only remote homologues were identified as potential template structures, no models have been built.
See here for template search results.
(61 residues)
>sp|P0DTC6|NS6_SARS2 MFHLVDFQVTIAEILLIIMRTFKVSIWNLDYIINLIIKNLSKSLTENKYSQLDEEQPMEI D
ORF6 protein (ORF6)
Disrupts bidirectional nucleocytoplasmic transport by interacting with the host RAE1-NUP98 complex (PubMed:33360543, PubMed:33849972). Disrupts cell nuclear import complex formation by tethering karyopherin alpha 2 and karyopherin beta 1 to the membrane (PubMed:32979938). Retention of import factors at the ER/Golgi membrane leads to a loss of transport into the nucleus (By similarity). Prevents STAT1 nuclear translocation in response to interferon signaling, thus blocking the expression of interferon stimulated genes (ISGs) that display multiple antiviral activities (PubMed:33097660). Suppresses IFN-beta production possibly by blocking IRF3 nuclear translocation (PubMed:32979938). Might induce accumulation of host HNRNPA1 (PubMed:33360543) May play a role in viral double membrane vesicles networks to enhance viral replication.
Experimental structures of hetero-oligomeric complexes (PDB: 7f60, 7vph) exist and high quality models can be extracted from them but should be used with care.
Only remote homologues were identified as potential template structures, no models have been built.
See here for template search results.
(121 residues)
>sp|P0DTC7|NS7A_SARS2 MKIILFLALITLATCELYHYQECVRGTTVLLKEPCSSGTYEGNSPFHPLADNKFALTCFS TQFAFACPDGVKHVYQLRARSVSPKLFIRQEEVQELYSPIFLIVAAIVFITLCFTLKRKT E
ORF7a protein (ORF7a)
Plays a role as antagonist of host tetherin (BST2), disrupting its antiviral effect (PubMed:33930332). Acts by binding to BST2 and sequestering it to perinuclear region, thereby preventing its antiviral function at cell membrane (PubMed:33930332). May specifically downregulate MHC-I allele HLA-A*02:01 (HLA-A2) (PubMed:36574644).
Partial coverage by experimental structures (PDB: 6w37, 7ci3) is available.
High quality models are available.
(43 residues)
>sp|P0DTD8|NS7B_SARS2 MIELSLIDFYLCFLAFLLFLVLIMLIIFWFSLELQDHNETCHA
ORF7b protein (ORF7b)
Only remote homologues were identified as potential template structures, no models have been built.
See here for template search results.
(121 residues)
>sp|P0DTC8|NS8_SARS2 MKFLVFLGIITTVAAFHQECSLQSCTQHQPYVVDDPCPIHFYSKWYIRVGARKSAPLIEL CVDEAGSKSPIQYIDIGNYTVSCLPFTINCQEPKLGSLVVRCSFYEDFLEYHDVRVVLDF I
ORF8 protein (ORF8)
Plays a role in modulating the host immune response (PubMed:31986261, PubMed:35343786, PubMed:36689483). May act as a secreted virokine by mimicking interleukin-17A (IL17A), and thereby binding to the IL17RA receptor, leading to activation of the IL17 pathway and increased secretion of pro-inflammatory factors (PubMed:35343786, PubMed:36689483). Contributes to the cytokine storm during SARS-CoV-2 infection when secreted by unconventional pathway (PubMed:33723527, PubMed:36689483). May act by down-regulating major histocompability complex class I (MHC-I) at cell surface (PubMed:34021074, PubMed:35157849). May inhibit expression of some members of the IFN-stimulated gene (ISG) family including hosts IGF2BP1/ZBP1, MX1 and MX2, and DHX58 (PubMed:34177923).
Experimental structures (PDB: 7f5f, 7jtl, 7jx6, 7mx9) are available. An experimental hetero-oligomeric complex (PDB: 7xmn) exists and a high quality model can be extracted but should be used with care.
High quality models are available which are predicted to form homo-dimers.
(419 residues)
>sp|P0DTC9|NCAP_SARS2 MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQHG KEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAG LPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGS QASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQLESKMSGKGQQ QQGQTVTKKSAAEASKKPRQKRTATKAYNVTQAFGRRGPEQTQGNFGDQELIRQGTDYKH WPQIAQFAPSASAFFGMSRIGMEVTPSGTWLTYTGAIKLDDKDPNFKDQVILLNKHIDAY KTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDDFSKQLQQSMSSADSTQA
Nucleoprotein (N)
Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M (PubMed:33264373). Plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication. Attenuates the stress granules formation by reducing host G3BP1 access to host mRNAs under stress conditions (PubMed:34901782, PubMed:36534661) May block host chemokine function in vivo, facilitating viral replication and transmission (PubMed:35921414). Acts by being secreted into the extracellular space where it competes to host chemokines for binding to host glycosaminoglycans (GAG) (PubMed:35921414) May induce inflammasome responses in cultured cells and mice. Acts by interacting with host NLRP3 to facilitate inflammasome assembly, which induces cytokine release that may play a role in COVID lung injury. Disrupts the association betwen NLRP3 and ABHD8, enhancing NLRP3 stability and leading to increased inflammasome activation (PubMed:39225180).
Experimental structures (PDB: 6m3m, 6vyo, 6wji, 6wkp, 6wzo, 6wzq, 6yi3, 6yun, 6zco, 7acs, 7act, 7c22, 7cdz, 7ce0, 7cr5, 7de1, 7f2b, 7f2e, 7ltu, 7lux, 7luz, 7lv2, 7n0i, 7n0r, 7n3c, 7n3d, 7o05, 7o35, 7o36, 7r98, 7sd4, 7str, 7sts, 7sue, 7uxx, 7uxz, 7vbd, 7vbe, 7vbf, 7vnu, 7xwx, 7xwz, 7xx1, 7xxk, 7ylb, 7yld, 8iqj, 8iv3, 8j6x, 8r6e, 8tfd, 8w6w, 8x1h, 8zbf, 8zfv, 9c2h, 9cj6, 9evy, 9ewh, 9exb, 9ezb, 9f2g, 9f2h, 9f2i, 9f5j, 9f5l, 9f7a, 9f7c, 9f83, 9fbg, 9in1, 9kn1, 9kur, 9qwi) are available. Partial coverage by experimental structures of hetero-oligomeric complexes exist (PDB: 7kgo, 7kgp, 7kgq, 7kgr, 7kgs, 7kgt, 7lg0, 7lgd, 7pku, 7qik, 7qip, 7suo, 7wzo, 7zit, 8dnt, 8th1, 8th5, 8utc, 8z05, 9f13, 9hlj, 9j4s, 9j4t, 9j4u, 9j4v, 9wbd) and high quality models can be extracted from them but should be used with care.
High quality models are available.
(97 residues)
>sp|P0DTD2|ORF9B_SARS2 MDPKISEMHPALRLVDPQIQLAVTRMENAVGRDQNNVGPKVYPIILRLGSPLSLNMARKT LNSLEDKAFQLTPIAVQMTKLATTEELPDEFVVVTVK
ORF9b protein (ORF9b)
Plays a role in inhibiting the host innate immune response by targeting the mitochondrial-associated innate immune response. Acts by binding to host TOMM70, inhibiting its binding to HSP90AB1 thereby disrupting the interferon activation pathway.
Experimental structures (PDB: 6z4u, 7ye7, 7ye8, 9mzb, 9n55) are available. Experimental structures of hetero-oligomeric complexes (PDB: 7dhg, 7kdt) exist and high quality models can be extracted from them but should be used with care.
High quality models are available which are predicted to form homo-dimers.
(73 residues)
>sp|P0DTD3|ORF9C_SARS2 MLQSCYNFLKEQHCQKASTQKGAEAAVKPLLVPHHVVATVQEIQLQAAVGELLLLEWLAM AVMLLLLCCCLTD
Putative ORF9c protein (ORF9c)
May induce apoptosis in cardiomyocytes when overexpressed ex-vivo.
Partial coverage by an experimental hetero-oligomeric complex (PDB: 7mb6) is available and a high quality model can be extracted but should be used with care.
Only remote homologues were identified as potential template structures, no models have been built.
See here for template search results.
(38 residues)
>sp|A0A663DJA2|ORF10_SARS2 MGYINVFAFPFTIYSLLLCRMNSRNYIAQVDVVNFNLT
Hetero-Oligomeric Complexes
Show sequences>sp|P0DTD1|R1AB_SARS2|3860-3942 SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLL SMQGAVDINKLCEEMLDNRATLQ
>sp|P0DTD1|R1AB_SARS2|3943-4140 AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLE KMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNII PLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNL AWPLIVTALRANSAVKLQ
nsp7 / nsp8 hetero-oligomeric complex
Nsp7-nsp8 hexadecamer may possibly confer processivity to the polymerase, maybe by binding to dsRNA or by producing primers utilized by the latter.
Experimental structures (PDB: 6m5i, 6wiq, 6wqd, 6wtc, 6xip, 6yhu, 7dcd, 7jlt) are available.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|3860-3942 SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLL SMQGAVDINKLCEEMLDNRATLQ
>sp|P0DTD1|R1AB_SARS2|3943-4140 AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLE KMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNII PLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNL AWPLIVTALRANSAVKLQ
>sp|P0DTD1|R1AB_SARS2|4393-5324 SADAQSFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNDKVAGFAKFLKTNCCRFQEKD EDDNLIDSYFVVKRHTFSNYQHEETIYNLLKDCPAVAKHDFFKFRIDGDMVPHISRQRLT KYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKKDWYDFVENPDILRVYANLGE RVRQALLKTVQFCDAMRNAGIVGVLTLDNQDLNGNWYDFGDFIQTTPGSGVPVVDSYYSL LMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERLKLFDRYFKYWDQTYHPNCVN CLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVN LHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAALTNNVAFQTVKPGNFNKDFY DFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYF DCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTIT QMNLKYAISAKNRARTVAGVSICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHN MLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLARKHTTCCSLSHRFYRLANECA QVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYV RNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSDDAVVCFNSTYASQGLVASIK NFKSVLYYQNNVFMSEAKCWTETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGA GCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADVFHLYLQYIRKLHDELTGHML DMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQ
nsp7 / nsp8 / Pol hetero-oligomeric complex
Experimental evidence for SARS-CoV that nsp7 and nsp8 activate and confer processivity to the RNA-synthesizing activity of Pol (Subissi et al., 2014 and Kirchdoerfer and Ward, 2019).
Experimental structures (PDB: 6m71, 6xqb, 6yyt, 7aap, 7b3b, 7b3c, 7b3d, 7btf, 7bv1, 7bv2, 7bw4, 7bzf, 7c2k, 7ctt, 7d4f, 7dfg, 7dfh, 7doi, 7dok, 7dte, 7krp, 7l1f, 7oyg, 7ozu, 7ozv, 7uo4, 7uo7, 7uo9, 7uob, 7uoe, 8gy6, 9blf, 9cgv, 9i81, 9l09, 9pyw, 9pyz, 9pz0, 9sao, 9sap, 9saq, 9sar) are available.
High quality models are available
>sp|P0DTD1|R1AB_SARS2|4254-4392 AGNATEVPANSTVLSFCAFAVDAAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPE ANMDQESFGGASCCLYCRCHIDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVC GMWKGYGCSCDQLREPMLQ
>sp|P0DTD1|R1AB_SARS2|5926-6452 AENVTGLFKDCSKVITGLHPTQAPTHLSVDTKFKTEGLCVDIPGIPKDMTYRRLISMMGF KMNYQVNGYPNMFITREEAIRHVRAWIGFDVEGCHATREAVGTNLPLQLGFSTGVNLVAV PTGYVDTPNNTDFSRVSAKPPPGDQFKHLIPLMYKGLPWNVVRIKIVQMLSDTLKNLSDR VVFVLWAHGFELTSMKYFVKIGPERTCCLCDRRATCFSTASDTYACWHHSIGFDYVYNPF MIDVQQWGFTGNLQSNHDLYCQVHGNAHVASCDAIMTRCLAVHECFVKRVDWTIEYPIIG DELKINAACRKVQHMVVKAALLADKFPVLHDIGNPKAIKCVPQADVEWKFYDAQPCSDKA YKIEELFYSYATHSDKFTDGVCLFWNCNVDRYPANSIVCRFDTRVLSNLNLPGCDGGSLY VNKHAFHTPAFDKSAFVNLKQLPFFYYSDSPCESHGKQVVSDIDYVPLKSATCITRCNLG GAVCRHHANEYRLYLDAYNMMISAGFSLWVYKQFDTYNLWNTFTRLQ
nsp10 / nsp14 hetero-oligomeric complex
Nsp10 plays a pivotal role in viral transcription by stimulating nsp14 3'-5' exoribonuclease activity.
Experimental structures (PDB: 7diy, 7mc5, 7mc6, 7n0b, 7n0c, 7n0d, 9fw2, 9fwh, 9fwi, 9fwj, 9fwk, 9fwl, 9fwm, 9fwn, 9fwo, 9fwp, 9fwq, 9fwr, 9fws, 9fwt, 9fwu, 9fz4, 9fzk, 9q1j, 9vck, 9vcl, 9yrk, 9yrl, 9yrn, 9yro) are available.
High quality models are available.
>sp|P0DTD1|R1AB_SARS2|4254-4392 AGNATEVPANSTVLSFCAFAVDAAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPE ANMDQESFGGASCCLYCRCHIDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVC GMWKGYGCSCDQLREPMLQ
>sp|P0DTD1|R1AB_SARS2|6799-7096 SSQAWQPGVAMPNLYKMQRMLLEKCDLQNYGDSATLPKGIMMNVAKYTQLCQYLNTLTLA VPYNMRVIHFGAGSDKGVAPGTAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVHT ANKWDLIISDMYDPKTKNVTKENDSKEGFFTYICGFIQQKLALGGSVAIKITEHSWNADL YKLMGHFAWWTAFVTNVNASSSEAFLIGCNYLGKPREQIDGYVMHANYIFWRNTNPIQLS SYSLFDMSKFPLKLRGTAVMSLKEGQINDMILSLLSKGRLIIRENNRVVISSDVLVNN
nsp10 / nsp16 hetero-oligomeric complex
Nsp10 plays a pivotal role in viral transcription by stimulating nsp16 2'-O-ribose methyltransferase activity.
Experimental structures (PDB: 6w4h, 6w61, 6w75, 6wjt, 6wkq, 6wks, 6wq3, 6wrz, 6wvn, 6xkm, 6yz1, 7bq7, 7c2i, 7c2j, 7jhe, 7jib, 7jpe, 7jyy, 7jz0, 7koa, 7l6r, 7l6t, 7lw3, 7lw4, 7r1t, 7r1u, 7ult, 8a23, 8bsd, 8bzv, 8c5m, 8f4s, 8f4y, 8osx, 8ot0, 8oto, 8otr, 8ov1, 8ov2, 8ov3, 8ov4, 8rv4, 8rv5, 8rv6, 8rv7, 8rv8, 8rv9, 8rva, 8rvb, 8rzc, 8rzd, 8rze, 8s8w, 8s8x, 8tyj, 8vuo, 9emj, 9eml, 9emv, 9eun, 9gny, 9grp, 9grq, 9gs4, 9gtf, 9gud, 9gue, 9guf, 9guy, 9gwo, 9p6p, 9q8h) are available.
High quality models are available.
(1273 residues)
>sp|P0DTC2|SPIKE_SARS2 MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFS NVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIV NNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLE GKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQT LLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETK CTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISN CVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIAD YNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPC NGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVN FNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITP GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSY ECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTI SVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQE VFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDC LGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALN TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPA ICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDP LQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDL QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDD SEPVLKGVKLHYT
(805 residues)
>sp|Q9BYF1|ACE2_HUMAN MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL NTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSGENP YASIDISKGENNPGFQNTDDVQTSF
Spike glycoprotein / ACE2_HUMAN hetero-oligomeric complex
Spike protein S1 binds to human ACE2, initiating the infection.
Experimental structures (PDB: 6lzg, 6m0j, 6vw1, 7a91, 7a92, 7a94, 7a95, 7a96, 7a97, 7a98, 7bh9, 7ct5, 7df4, 7dmu, 7dqa, 7dx4, 7dx5, 7dx6, 7dx7, 7dx8, 7dx9, 7edj, 7efp, 7efr, 7ekc, 7eke, 7ekf, 7ekg, 7ekh, 7fdg, 7fdh, 7fdi, 7fem, 7kj2, 7kj3, 7kj4, 7kmb, 7kms, 7kmz, 7knb, 7kne, 7knh, 7kni, 7l0n, 7l7f, 7lo4, 7mjm, 7mjn, 7nxc, 7p19, 7r0z, 7r10, 7r11, 7r12, 7r1a, 7rpv, 7sn0, 7sxx, 7sxy, 7sxz, 7sy0, 7sy1, 7sy2, 7sy3, 7sy4, 7sy5, 7sy6, 7sy7, 7sy8, 7t9k, 7t9l, 7tew, 7tex, 7tez, 7tf0, 7tn0, 7u0n, 7ufk, 7ufl, 7v7z, 7v80, 7v81, 7v82, 7v83, 7v84, 7v85, 7v86, 7v87, 7v88, 7v89, 7v8a, 7v8b, 7vx4, 7vx5, 7vx9, 7vxa, 7vxb, 7vxc, 7vxd, 7vxf, 7vxk, 7vxm, 7w99, 7w9b, 7w9c, 7wbl, 7wbp, 7wbq, 7wgb, 7wgc, 7whh, 7wk4, 7wk5, 7wk6, 7wnm, 7wpa, 7wpb, 7wpc, 7ws8, 7ws9, 7wsa, 7wvp, 7wvq, 7xaz, 7xb0, 7xb1, 7xch, 7xci, 7xcp, 7xid, 7xo7, 7xo8, 7xo9, 7xwa, 7y1y, 7y1z, 7y20, 7y21, 7y9z, 7ya0, 7ya1, 7ydi, 7yeg, 7yhw, 7yj3, 7yr2, 7yr3, 7yr4, 7zdq, 7zf7, 8aqs, 8df5, 8dlj, 8dlk, 8dlm, 8dln, 8dlp, 8dlq, 8dlu, 8dlv, 8dm5, 8dm6, 8dv1, 8dv2, 8h06, 8h5c, 8hrk, 8hrl, 8i9b, 8i9c, 8i9d, 8i9e, 8i9f, 8i9g, 8i9h, 8if2, 8iou, 8iov, 8jje, 8jyn, 8jyo, 8jyp, 8qsq, 8sph, 8spi, 8vko, 8vkp, 8vqr, 8wdr, 8wds, 8wdy, 8wdz, 8we0, 8we1, 8we4, 8wgv, 8wgw, 8whs, 8whu, 8whz, 8wp8, 8wrh, 8wrl, 8wrm, 8wro, 8wtd, 8wtj, 8wyh, 8xal, 8xlm, 8xln, 8xn2, 8xn3, 8xn5, 8xnf, 8xnk, 8xsf, 8xsj, 8xuy, 8xuz, 8xv0, 8xv1, 8xvm, 8xxw, 8xy9, 8xye, 8xyg, 8xyo, 8y16, 8y18, 8y6a, 8yzb, 8yzc, 8yzd, 8yze, 8z3w, 8z4x, 8z64, 8z6a, 8z7b, 8z7p, 8zbq, 9bnd, 9bne, 9bnf, 9bng, 9ele, 9elf, 9iu1, 9iup, 9iuq, 9iuu, 9ue6, 9ue7) are available.
High quality models are available.
Structural Analysis of Mutations
We downloaded SARS-CoV-2 mutation data from GISAID (July 3, 2021) and computed the frequencies of every observed mutation. We summarized the number of different mutations observed at each residue position and summed the frequencies for each residue. For insertions and deletions the frequencies were assigned to the previous residue, except for N-terminal insertions which were assigned to the first residue of the protein.
We performed proximity searches within an 8A radius, and summed both frequencies and number of mutations (excluding the residue itself). To normalize the mutations we first computed the residue-level average of frequencies and number of mutations for each UniProt protein. We then subtracted this baseline value from each residue in the proximity that was mapped to a SARS-CoV-2 protein by SIFTS.
| Residue-level analysis | Proximity (8A) | Proximity (8A, normalized) | |
|---|---|---|---|
| Number of mutation | View results | View results | View results |
| Number of mutation (in binding sites) | View results | View results | View results |
| Mutation frequency | View results | View results | View results |
| Mutation frequency (in binding sites) | View results | View results | View results |
Click on a project to list its content on the SWISS-MODEL Repository.
We gratefully acknowledge the authors, originating and submitting laboratories of the genetic sequence
and metadata made available through GISAID on which this research is based.
You can download the
full list of sequence IDs, submission date and location used in the analysis here.
If you use this data in a publication, please cite: Elbe S., and Buckland-Merrett G. (2017)
"Data, disease and diplomacy: GISAID’s innovative contribution to global health." Global Challenges 1: 33-46
![]()
![]()
Part of the analysis was performed by collaborators at the Computational Biology group of BSC-CNS within the EXSCALATE4CoV
project.